-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Virology and Molecular Biology
p-ISSN: 2163-2219 e-ISSN: 2163-2227
2016; 5(1): 1-7
doi:10.5923/j.ijvmb.20160501.01

Characterization of Subviral Particles of Hepatitis B Virus Produced by HepG2.2.15 Cell Line - In vitro Study
Mohamed M. S. Farag 1, Mohamed T. M. Mansour 2
1Botany and Microbiology Department, Faculty of Science, Al-Azhar University, Cairo, Egypt
2Children’s Cancer Hospital, Cairo, Egypt
Correspondence to: Mohamed M. S. Farag , Botany and Microbiology Department, Faculty of Science, Al-Azhar University, Cairo, Egypt.
| Email: |  |
Copyright © 2016 Scientific & Academic Publishing. All Rights Reserved.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Hepatitis B virus (HBV) is unusual in that its surface proteins small, medium, and large are not only incorporated into the virion envelope but they also bud into empty subviral particles (SVP) in great excess over virions. In the sera of patients infected with HBV, in addition to infectious particles, there is an excess (typically 1,000- to 100,000-fold) of empty SVP. The main goal of this study was to characterize these subviral particles that released from integrated HepG2.2.15 cell line in vitro. To achieve this aim, integrated HepG2.2.15 cell line was cultivated for production and concentration of SVP. The SVP were examined by serological markers, molecular assay, western blot, and electron microscopy. Our results indicated that cultivation of HepG2.2.15 cell line in complete williams medium E including insulin and hydrocortisone results in production high amounts of secreted viral particles in the supernatant. The SVP showed both surface and core proteins sharing the same HBV virion proteins. Molecular assay indicated that SVP was empty particles and didn’t contain full genome. For the morphogenesis and structures of SVP, electron microscopy showed different spherical and filaments shapes for SVP sharing the same morphology of HBV virion. These findings shed light on an important step in the HBV infectious cycle, as the intracellular accumulation of HBV subviral particles may be directly linked to viral pathogenesis.
Keywords: HBV, SVP, HepG2.2.15 cell line
Cite this paper: Mohamed M. S. Farag , Mohamed T. M. Mansour , Characterization of Subviral Particles of Hepatitis B Virus Produced by HepG2.2.15 Cell Line - In vitro Study, International Journal of Virology and Molecular Biology, Vol. 5 No. 1, 2016, pp. 1-7. doi: 10.5923/j.ijvmb.20160501.01.
Article Outline
1. Introduction
- The human hepatitis B virus (HBV) is an enveloped DNA virus and the prototype of the Hepadnaviridae family. HBV is characterized by a strict tropism for human hepatocytes and the ability to cause acute and chronic infections [10]. Chronic infection with HBV affects 350 to 400 million individuals worldwide and is the leading cause of liver cirrhosis and hepatocellular carcinoma (HCC) worldwide [17, 22]. More than 780,000 people die every year due to the consequences of hepatitis B (WHO 2014). The virion (or Dane particle) is a spherical particle, 42 nm in diameter, consisting of an icosahedral nucleocapsid of approximately 30 nm in diameter and an envelope composed of three surface proteins and presumably lipids of host cell origin. The nucleocapsid and envelope are synthesized and mature separately in different cellular compartments, subsequently interacting to form the virion [4, 18, 25]. HBV hepatotropism is determined, for the most part, by the HBV envelope proteins at viral entry. A single open reading frame in the HBV genome encodes three envelope proteins that differ from each other by the size of their N-terminal ectodomain. They bear the HBV surface antigen (HBsAg) in their common S domain and are referred to as the large HBsAg (L-HBsAg), middle HBsAg (M-HBsAg), and small HBsAg (S-HBsAg) proteins. They are produced in quantities far exceeding the amount needed for the assembly of HBV virions [4, 15, 24], and due to their capacity for self-assembly, they are secreted abundantly as empty subviral particles (SVP).In infections with HBV, there is an excess of empty noninfectious SVP that do not contain the viral capsid. The SVP are typically present in a 1,000- to 100,000-fold excess relative to the infectious particles [10, 19]. The SVP are spherical and filamentous particles, 22-25 nm in diameter [12, 14, 30], with an octahedral symmetry [26], lack the nucleocapsid, does not contain the viral genome, and is not infectious [27]. As compared to the virion, the subviral particles are highly over produced. They can contain all three forms of the HBV envelope proteins: L, M, and S. These share a common C terminus, with M containing the pre-S2 domain relative to S and L containing the pre-S1 domain relative to M [19]. There is good evidence that during infection a domain within the pre-S1 of L is what interacts with as-yet-unidentified host receptors [12]. The SVP from patients are immunogenic and were used with success in the first HBV vaccine [1]. Most current available HBV vaccines are prepared in yeast from SVP assembled using just the HBV S protein; such SVP are sufficient to protect individuals against HBV and HDV.The biological function of this excess of SVP has been largely ignored. Some authors have suggested that SVP might stop up neutralizing antibodies produced by the host and thus increase the ability of the infectious particles to reach susceptible cells [9, 28]. It has also been suggested that SVP contribute to a state of immune tolerance that is a precondition for highly productive persistent infection [11].HBsAg is secreted from infected hepatocytes as SVP, which can accumulate up to concentrations of 100 μg/ml in peripheral blood [2]. During the infection, it is currently assumed that these SVP act as decoys, trapping the host’s immune system to protect the infectious virus [23]. The high amounts of HBsAg in the circulation of patients can theoretically contribute to the hampered immune response [2]. Interestingly, it has been clearly shown that this natural phenomenon specific to HBV is linked to the ability of the SHBs envelope protein to spontaneously form empty envelope particles at the rough endoplasmic reticulum (ER) 1. This noninfectious particle was the basis of most vaccines against hepatitis B [23].As described below, we assembled different forms of SVP that released from integrated HepG2.2.15 cell line in vitro. The SVP were examined by serological markers, molecular assay, western blot, and electron microscopy. These findings shed light on an important step in the HBV infectious cycle, as the intracellular accumulation of HBV subviral filaments may be directly linked to viral pathogenesis.
2. Materials and Methods
2.1. HepG2.2.15 Cell Line and Culture Condition
- The human hepatoma cell line HepG2.2.15 contains a genomic integrated HBV genome were cultivated in tissue culture flasks in 10 ml of complete Williams medium E (Lonza, Swiss) including 10% (v/v) heat-inactivated fetal calf serum (FCS) (Serolab, Brazil), 100 U of penicillin/ml, 100 μg of streptomycin/ml (Lonza, Swiss), 2 mM L-glutamine (Lonza, Swiss), 250 µl insulin (5µg/ml) (Sigma, China), and 250 µl hydrocortisone (Sigma, China). The cells were cultivated at 37°C in a CO2 incubator. After 90% cell confluence, the supernatant was harvested and collected in large falcon tubes for virus concentration. For cell trypsination, passaging of adherent cells were performed using trypsin/EDTA (Lonza, Swiss) at 37°C. Prior to trypsination the cells were carefully washed with PBS buffer to remove cell culture medium. After 2-5 minutes the treatment was stopped by addition of 10 mL culture medium including FCS. If necessary, cell aggregates were minimized by gently pipetting up and down to suspend the cells. The cells were seeded in fresh culture medium to reach 30-50% confluency.
2.2. Optimizing the Production of SVP
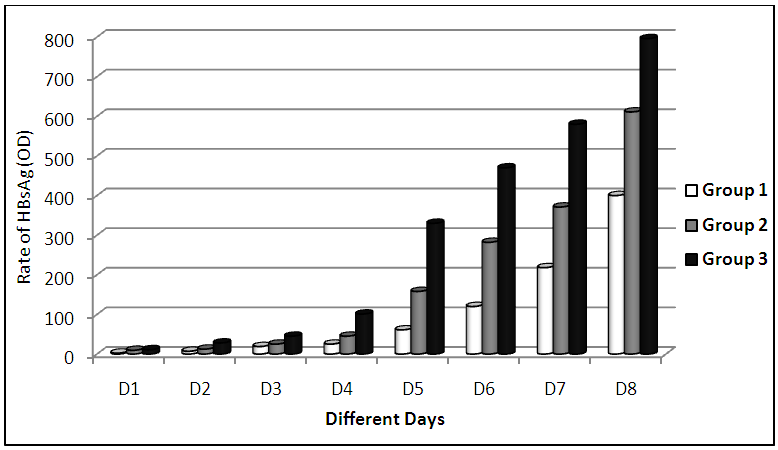
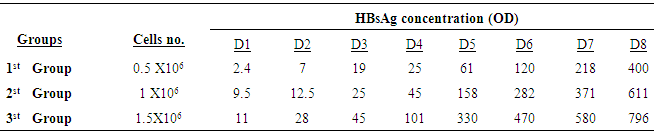
- To optimize the production of SVP, the commercially available human hepatoma cell line HepG2.2.15 containng a genomic integrated HBV genome was used. These cells continuously produce and express HBVsvp into the supernatant. The SVP was detected in the supernatant by ELISA for the secreted surface antigen (HBsAg) according to manufacturer instructions. The HepG2.2.15 was cultivated at flask 75 cm2 with 20 ml complete William's E media with different cell number distributed at 3 groups (each group consist of 5 flasks). Cell numbers was (0.5 X 106) for the 1st group. Regarding to the 2nd group the count of cells was (1 X 106). Finally, the count of cells was (1.5 X 106) for the 3rd group. HBsAg was detected by ELISA (Germany) at day 1, 2, 3, 4, 5, 6, 7, and 8 for optimizing virus production.
2.3. HBVsvp Concentration
- HBVsvp were prepared and concentrated by the polyethylene glycol protocol, which required the harvested medium to be incubated overnight at 4°C with polyethylene glycol (40%) (Nice, India). On day two, the medium was centrifuged at 8000 rpm for one hour at 4°C. After centrifugation, the pellet was resuspended in 1x PBS (75%) containing FCS (25%), followed by incubation for a second night at 4°C. On day three, the supernatant (isolates) were harvested, and centrifuged again at 4000 rpm for 20 minutes at 4°C. After centrifugation the isolates were collected and stored at -80°C.
2.4. Molecular Assay
- For DNA extraction, viral DNA was extracted according to the manufacturer’s instructions using Ribo virus DNA extraction kit (Sacace, Italy). A 465 bp HBX gene fragment was amplified using PCR. HBX gene was amplified using forward 5´ATGCAAGCTTA TGGCTGCTAGGCTGTACTG-3´and reverse5´TGCGAATTCTTAGGCAGAGGTGAAA AAGTTG-3´ primers. The PCR amplification was performed at 95°C for 5 min followed by 35 cycles at 94°C for 1 min, 61°C for 45 s and 72°C for 1 min. Final extension was performed at 72°C for 10 min. The amplification was performed in programmable thermal cycler (Eppendorf, Germany). The PCR products were visualized by electrophoresis using ethidum bromide in 2% agarose gel (Gibco-BRL, Life technology, MD, USA).
2.5. Western Blotting
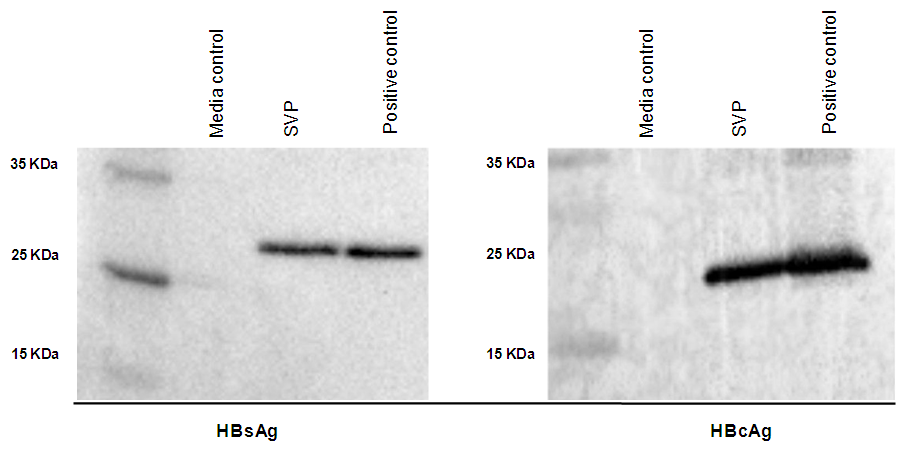
- The SVP of hepatitis B virus that produced by HepG2.2.15 cell line was investigated for different surface and core proteins. The cells were lysed in lysis buffer containing 150 mM NaCl, 1% SDS, 1% NP-40 and 0.5% sodium deoxycholate. The concentrated supernatant of cell lysates was run on 12% SDS-polyacrylamide gel and electrophoretically transferred to nitrocellulose membranes. After blocking, membranes were incubated with primary antibodies against HBs protein and HBc protein at 4°C overnight. After incubation with secondary antibody, bands were detected western blotting detection system.
2.6. Electron Microscopy
- Different samples from chronic HBV patients and SVP produced HepG2.2.15 cell line were examined and characterized by transmission electron microscopy (JEOL 1010, Japan) at a voltage of 200 KV to know the size and shape of the particles. The samples were prepared by drop-coating the samples solution onto the carbon-coated copper grid, dried under infrared lamp, and were loaded onto a specimen holder and analyzed by transmission electron microscopy.
3. Results
3.1. Virus Production and Optimization
- The cultivation of HepG2.2.15 in complete Williams medium E including insulin and hydrocortisone results in continuous production and expression high amounts of secreted SVP in the supernatant. Figure 1 showed the morphology of HepG2.2.15 cell line at different cell count groups; A) 0.5 X 106 for the 1st group, B) 1 X 106 for the 2nd group, and finally C) 1.5 X 106 for the 3rd group. Our results indicated that the highest virus secretion was found at day 8 of splitting cells in each different cell count group. The titre of virus confirmed by detection of HBsAg was 400, 611, 796 (OD) for first, second, and third groups, respectively. The second day for the best virus production was day 7, the titre of virus was 218, 371,580 (OD) for first, second, and third groups, respectively. The third day for the best virus production was day 6, the titre of virus was 120, 282,470 (OD) for first, second, and third groups, respectively (Table 1; Figure 2). The lowest virus titre was at day 1, the titre of virus was 2.4, 9.5, 11 (OD) for first, second, and third groups, respectively (Table 1; Figure 2).
|
3.2. SVP Preparation and Concentration
- For this assay the HepG2.2.15 cells were propagated in several runs. The SVP was concentrated by the polyethylene glycol protocol. The media were collected and HBsAg was assayed before and after SVP concentration (Table 2; Figure 3). We were able to concentrate the SVP in very high titers. Each run was done in this experiment was average for different collected supernatant. The highest SVP concentrations were 2785, 2626 (OD) for runs 1 and 3, respectively. The lowest SVP concentration was 971 (OD) for run 2 (Table 2; Figure 3).
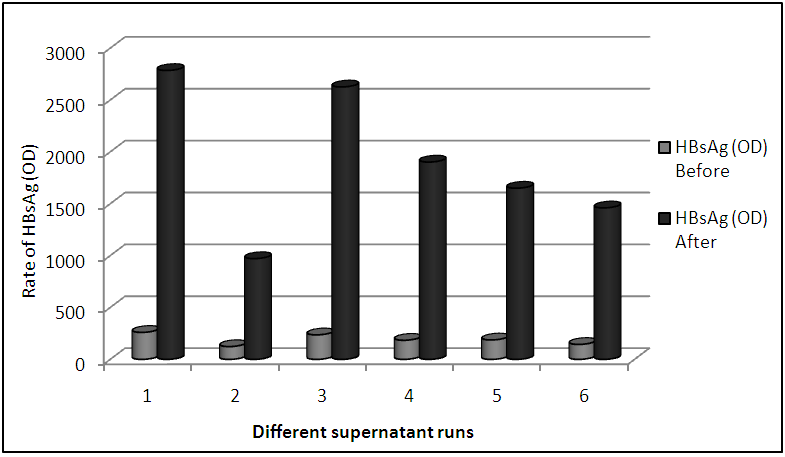 | Figure 3. HBsAg was measured by ELISA before and after SVP concentration for different supernatant runs |
|
3.3. Detection and Analysis of DNA
- For HBX gene detection, SVP produced by HepG2.2.15 cell line was analysed for the presence of HBX gene using a specific primers. Our results showed that SVP was empty particles because a 465 bp fragment of HBX gene didn’t detected in our sample (Figure 4).
 | Figure 4. Confirmation the absence of 465 bp fragment of HBX gene in SVP sample, which indicate that SVP was empty particles |
3.4. Assay of HBV-specific Proteins by Western Blotting Analysis
- The SVP was subjected to western blot analysis using antibodies against HBsAg and HBcAg showed both surface and core proteins sharing the same HBV virion proteins. Our results indicated that both HBs and HBc proteins were detected for SVP harvested from supernatant of HepG2.2.15 cell line, 24 kDa small surface antigens, and 21 kDa for core protein comparing to provided controls (Figure 5).
3.5. Examination of HBVsvp by Electron Microscopy
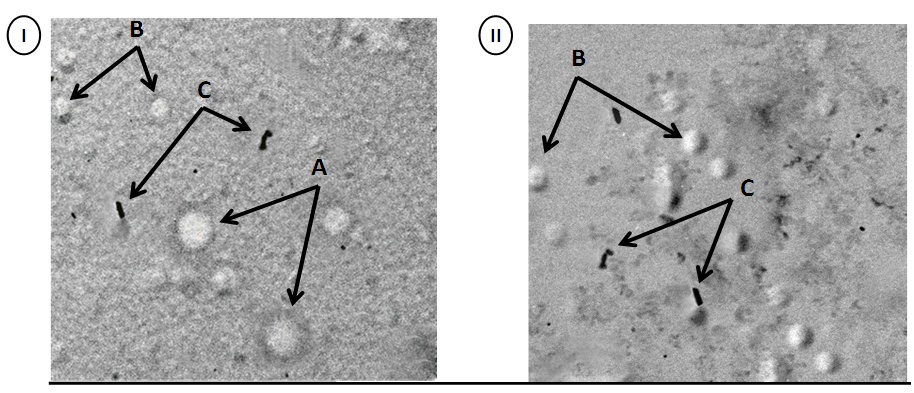
- To differentiate between Dane-like particles and subviral particles of HBV, positive samples and supernatant of HepG2.2.15 cell line were examined by transmission electron microscopy. Our results showed that positive HBV samples showed both HBV (Dane-like particle) and HBVsvp. Regards to supernatant of HepG2.2.15 cell line, SVP of HBV only was detected. The samples examined by electron microscopy showed A) = 42 nm HBV (Dane-like particle), B) = 22 nm spherical subviral particle, C) = 22 nm filamentous subviral particle (Figure 6).
4. Discussion
- HBV contains three coterminal envelope proteins on the virion surface: large (L), middle (M), and small (S) [32]. The L, M, and S proteins are also secreted as empty “subviral particles,” which exceed virions by at least 1,000-fold. The S protein serves as the morphogenic factor for both types of particles, while the L protein is required only for virion formation [32]. Of the three envelope proteins produced both by HBV particles, the L and S proteins are essential for virion secretion, whereas the M protein is dispensable [6]. The major objective of the present study was to characterize these subviral particles that released from integrated HepG2.2.15 cell line in vitro. The establishment of an in vitro system in which the replication, pathobiology, and oncogenic potential of HBV can be investigated has proven to be elusive. The advantage of this system over the system reported is that the penetration and the uncoating of the virus can be studied in agreements with previous report [25]. The replication in this system occurs continuously for an extended time period, which is a not feasible using primary liver cell [25]. In the present study HBVsvp were characterized for several reasons. The first reason is that they are not infectious and can therefore be considered safe in handling in agreement with previous investigations [10, 12, 14, 19, 29, 30]. Although there are infectious particles found in the HepG2.2.15 supernatant, they are at very low concentration (1:100.000–1.000.000) and could be completely cleared. In contrast, other results suggest that the hepatoma cell lines can produce infectious virus after transfection with the viral genome [13, 31]. Another important reason for choosing HBVsvp for characterization was that they consist of the two major structural proteins of HBV and present these proteins in a natural fashion, sharing the same HBV virion proteins. We consider these SVP particles isolated from HepG2.2.15 are more defined than SVP obtained from the sera of infected individuals: the latter contain infectious virus as well as SVP, they may also contain a spectrum of host antibodies either mixed with the SVP or even directly attached to the SVP, and since the patients producing SVP are chronically infected, the genetic composition of the SVP will be mixed, with a variety of mutant forms [7]. Taken together, there are valuable reasons to characterize HBVsvp that they are available by HepG2.2.15 in almost unlimited amounts and additionally, they could be produced for the different genotypes and subspecies. These are important points, since it would offer the possibility to vaccinate people prior to infection, for example non-responders to the available vaccine. We have demonstrated that the cultivation of HepG2.2.15 in complete williams medium E including insulin and hydrocortisone results in continuous production and expression high amounts of secreted SVP in the supernatant. On the other hand, these particles provide a model to develop therapeutic vaccines against HBV infection in agreement with previous investigations [8, 16, 21].Despite the existence of hepatoma cell lines that can produce infectious virus after transfection with the viral genome [13, 31] and the recent establishment of a cell line that can be infected by the virus in vitro [13], little is known about the morphogenesis of HBV. This may be due to the low rate of HBV production in these cells, as it has been estimated that an individual hepatocyte releases only 1 to 10 viruses per day during the productive phase of infection in vivo [20]. HBV subviral envelope particles can reach concentrations 10,000 times higher than that of virions in the serum of infected patients, but their assembly and intracellular transport also remain poorly understood [5]. As the production of the S envelope protein alone is sufficient for the secretion of spherical HBV subviral envelope particles, it is widely thought that the filamentous shape of these particles results from the coproduction of the S and L proteins [3, 25]. Our electron microscopy results showed that HepG2.2.15 cell line integrated with HBV genome was able to produce spherical and filamentous SVP. Assembling of SVP from HepG2.2.15 provided some interesting insights. (i) Under our experimental conditions, there were excess folds of SVP produced. (ii) These SVP contained no detectable DNA. (iii) SVP envelope proteins were 22-25-nm spherical to filaments particles. (iv) The involvement of the immune system in the pathogenesis of hepatitis is well established but the mechanism for this interaction has not yet been clarified. The transfected HepG2.2.15 cells have the potential to serve as an in vitro model system for the investigation of the interaction of specific cytotoxic T lymphocytes and cytolytic antibodies with infected liver cells. In conclusion, we have demonstrated that the cultivation of HepG2.2.15 in complete williams medium E including insulin and hydrocortisone results in continuous production and expression high amounts of secreted SVP in the supernatant. The SVP showed both surface and core proteins sharing the same HBV virion proteins. For the morphogenesis and structures of SVP, electron microscopy showed different spherical and filaments shapes for SVP sharing the same morphology of HBV virion.These findings shed light on an important step in the HBV infectious cycle, as the intracellular accumulation of HBV subviral particles may be directly linked to viral pathogenesis and provided a model for prophylactic and therapeutic vaccine against HBV infection.
ACKNOWLEDGEMENTS
- We thank Professor Dr Rolf Bartenschlager, Department of Molecular Virology, University of Heidelberg and his group for their kind gift of HepG2.2.15 integrated cell line.I would like to express my deepest gratitude to ministry of state for scientific research in Egypt and Science and technology development fund (STDF) for financing this article.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML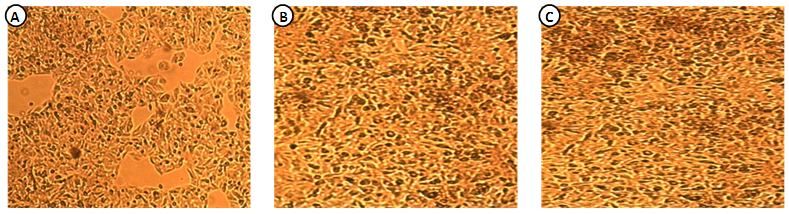
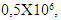
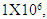 and
and 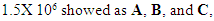 respectively
respectively