-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Virology and Molecular Biology
p-ISSN: 2163-2219 e-ISSN: 2163-2227
2013; 2(1): 1-6
doi:10.5923/j.ijvmb.20130201.01
Distribution of Chlamydia trachomatis Genotypes in Infertile Patients of Córdoba, Argentina
Monetti M. S.1, Molina R.2, Estofan P.3, Frutos M. C.1, Kiguen A. X.1, Venezuela R. F.1, Paglini G.1, Cuffini C.1, 4
1Institute of Virology, National University of Cordoba. Cordoba, 5016, Argentine
2Laboratory of Reproductive Andrology. Córdoba, Argentina
3Integral Center of Ginecology, Obstetric and Reproduction. Córdoba, Argentina
4Fleming 3498, Barrio: Lago Azul Villa Santa Cruz del Lago Córdoba, 5152, Argentine
Correspondence to: Cuffini C., Institute of Virology, National University of Cordoba. Cordoba, 5016, Argentine.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
To detect and characterize Chlamydia trachomatis (C. trachomatis) genotypes in infertile patients of Córdoba, Argentina; 660 endocervical and urethral swabs and semen samples were collected from infertile patients for detection of C. trachomatis by omp A gene with Hemi Nested-PCR and cryptic plasmid-PCR. Sequencing methods of omp A gene were used to identify C. trachomatis genotypes. The sequences obtained were aligned with chlamydial sequences currently available in the GenBank, for the design of the phylogenetic tree. The prevalence of C. trachomatis was 7.27% (48/660). We did not detect C. trachomatis cryptic plasmid free strains. According to the results of nucleotide sequences, the distribution of genotypes was L1 (50 %) followed by G (25 %), E (12.5%) and D (12.5%). Patients who tested positive to genotype L1 had no symptoms of lymphogranuloma venereum (LGV). This is the first study that provides information about the distribution of C. trachomatis genotypes and the circulation of cryptic plasmid negative strains of C. trachomatis among patients with infertility in Córdoba, Argentina.
Keywords: Chlamydia trachomatis, Genotypes, Infertile Patients
Cite this paper: Monetti M. S., Molina R., Estofan P., Frutos M. C., Kiguen A. X., Venezuela R. F., Paglini G., Cuffini C., Distribution of Chlamydia trachomatis Genotypes in Infertile Patients of Córdoba, Argentina, International Journal of Virology and Molecular Biology, Vol. 2 No. 1, 2013, pp. 1-6. doi: 10.5923/j.ijvmb.20130201.01.
Article Outline
1. Introduction
- Chlamydia trachomatis (C. trachomatis) is the most prevalent bacteria in sexually transmitted infections (STI) and can result in severe genital and ocular diseases[8]. The WHO has estimated 100 annual million new cases worldwide; however, most of the women with lower genital tract infections remain asymptomatic and therefore, undiagnosed[17].The primary sites of C. trachomatis infections are female endocervix and urethra of both genders. In men, C. trachomatis is associated with non-gonococcal urethritis and epididymitis[6]; in women, this pathogen can lead to serious complications such as endometritis, salpingitis, pelvic inflammatory disease, ectopic pregnancy or tubal factor infertility[1, 26]. However, many patients remain asymptomatic and develop persistent infections, which can lead to severe reproductive sequelae[13]. Infertility due to C. trachomatis represents a preventable disease when is detected early on[21].C. trachomatis possesses a cryptic 7.5-kb plasmid of unknown function that is a preferred target for various nucleic acid amplification tests, since it contains multiple copies. However, among C. trachomatis strains, 22 plasmid free variants have been described; therefore, it is possible to obtain false negative results when cryptic plasmid PCR is the only test used [20].C. trachomatis strains are currently classified into genotypes using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) or sequencing of the ompA gene, which encodes the major outer membrane protein (MOMP). Genotypes A-C cause primary trachoma/blindness, D–K produce urogenital infections and L1–3 are responsible for invasive lymphogranuloma venereum (LGV)[15]. Genotypic characterization of C. trachomatis isolates can not only provide valuable insights into circulating C. trachomatis genotypes within a given community, but also improve understanding of their epidemiology, which may assist in developing new strategies for improving STI control[4, 12].In Argentina, the distribution of genotypes in patients with infertility is still unknown. The aim of this study was to detect and characterize Chlamydia trachomatis (C. trachomatis) genotypes in patients with infertility in Córdoba, Argentina. We consider that this study is important to contact tracing and monitoring, to enable associations with clinical manifestations or pathogenicity and that it may also play a role in developing strategies for vaccine design.
2. Materials and Methods
2.1. Clinical Samples
- Six hundred and sixty urogenital specimens were collected from adult patients (439 women and 221 men; mean age: 36.7 years[r=21-55]) who consulted for infertility at private breeding centers of Cordoba city, Argentina, between January and July, 2012. Samples were classified in: cervical swabs (CS) (n: 437), urethral swabs (US) (n: 5), urethral swabs with semen (US+S) (n: 148) and semen samples (n: 70). All of them were obtained by health care professionals and placed in sterile tubes containing 1 ml of SPG (sucrose, phosphate, glutamic acid), and subsequently sent to the Instituto de Virología, Facultad de Ciencias Médicas, Universidad Nacional de Córdoba, Argentina.Inclusion criteria: adult patients of both genders who complained of impaired fertility. Exclusion criteria: pregnant women, immunocompromised patients.The project was approved by the Ethics Committee C.E.I.E.S. Oulton-Romagosa, Córdoba and all patients signed written informed consent before entering the study.
2.2. DNA Extraction
- 200 μl of each sample were subjected to DNA extraction using the Accuprep Genomic DNA Extraction Kit (BIONEER, Alameda, CA, USA) according to the manufacturer’s instructions.
2.3. OmpA gene Hemi Nested PCR
- PCR DNA extract (5 μl) was used to amplify a 1045 pb fragment of the ompA gene of C. trachomatis, using primers SeroA1 (5’ATGAAAAAACTCTTGAAATCGG3’) and SeroA2 (5’TTTCTAGAT/ CTTCATT/CTTGTT3´); in the nested PCR, it was replaced by the first SeroA1 by PCTM3 (5’TCCTTGCAAGCTCTGCCTGTGGGGAATCCT3’). Both PCR amplification processes commenced with a 4-minute denaturation step at 95°C and continued with 49 amplification cycles. Each cycle consisted of a first denaturation step at 95°C for 1 min, an annealing step at 55°C for 1 min and a final step of chain elongation at 72° C for 1.5 min.[16].
2.4. Cryptic Plasmid PCR
- The primers used to generate a 201-bp fragment from the cryptic plasmid of C. trachomatis were CTP1(5'-TAGTAACTGCCAClTCATCA-3') and CTP2(5'-TTCCCCTTGTAATTCGTTGC-3'). The PCR amplification consisted of DNA denaturation at 95°C for 4 min followed by 35 cycles of amplification with a thermocycler Model One, Germany. Each cycle consisted of 1 min at 95°C, 1 min at 55°C and 1.5 min at 72°C followed by a final elongation at 72°C for 4 min. The ompA gene and cryptic plasmid PCR products were visualized after electrophoresis in a 1% agarose gel by ethidium bromide staining[16].
2.5. Sequencing of the omp A gene
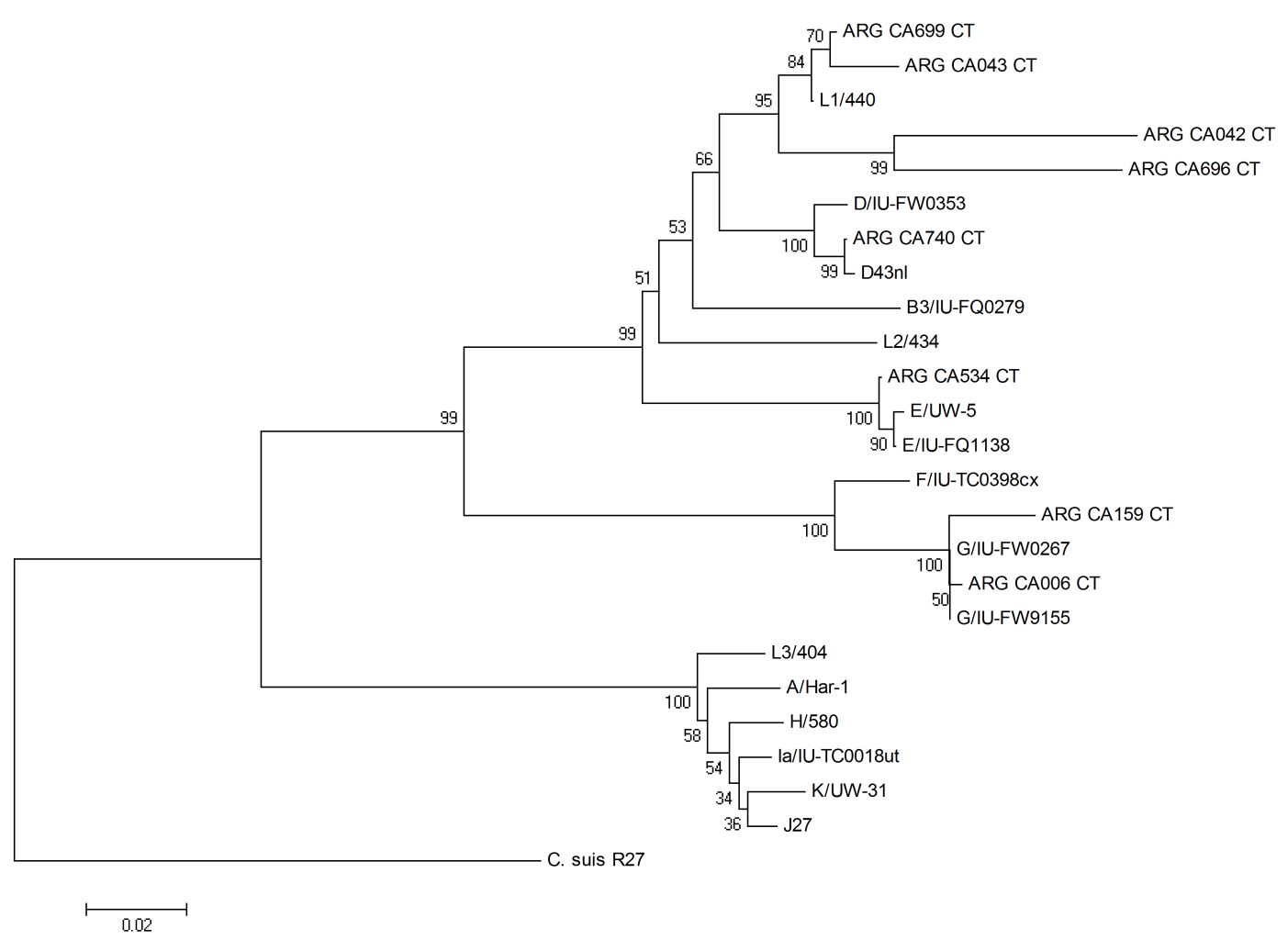
- For sequence analysis, the nested-PCR products were purified with the QIAquick Gel Extraction Kit (Qiagen, Valencia, CA, US) and subjected to direct nucleotide sequencing reaction in both directions using an ABI automatic sequencer. The sequences were analyzed using the Molecular Evolutionary Genetics Analysis software package, MEGA4[25]. Sequences of the ompA derived from strains used in this study were analyzed along with the next sequences from strains available in GenBank: F/IU-IC0398cx (accession number FJ261947.1), E/UW-5 (HQ637270.1), E/IU-FQ1138 (FJ261931.1), G/IU-FW0267 (FJ261928.1), G/IU-TC0398cx (FJ261947.1), L1/440 (DQ064294.1), L2/434 (DQ064295.1), L3/404 (DQ064296.1), D/IU-FW0353 (FJ261929.1), D43nl (JN795446.1), K/UW-31 (DQ064293.1), H/580 (DQ064289.1), Ia/IU-IC0018ut (FJ261940.1), J-27 (JN795448.1), A/Har-1 (DQ064279), B3/IU-FQ0279 (FJ261925.1) and the tree was rooted with the ompA sequence of the Chlamydia suis (C. suis) strain (accession number AF26273.1). Phylogenetic tree was constructed using the neighbor-joining method[24]. Branching pattern confidence levels were estimated by the bootstrap resampling of the data based on 1000 random replicates.
2.6. Statistical Analysis
- Statistical analysis was performed using Chi-square or Fisher exact test. P value lower than 0.05 was considered statistically significant. Absolute and relative frequencies and 95% confidence intervals (CI) were given.
3. Results
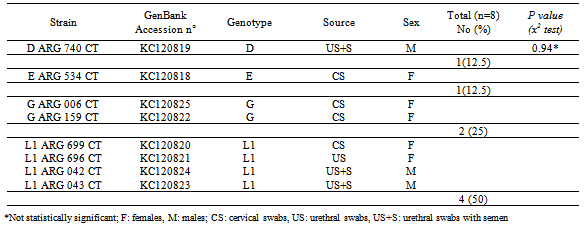
- C. trachomatis cryptic plasmid was detected by PCR in 48/660 of the cases (7.27%), the prevalence was 7.52% (33/439) in women and 6.79% (15/221) in men. The difference was statistically significant p= 0.0003, OR= 3.81 (1.67-8.96, CI 95%). The mean age (years±SD) for infected men and women was 36.7±1.5 and 35.4±4.5, respectively. The percentage of detection in CS was 66.67% (32/48), while in US+S was 18.75% (9/48), only in semen 12.5% (6/48) and only in US 2.08% (1/48), being significantly higher (p<0.001) in samples from cervical swabs. In this study, C. trachomatis cryptic plasmid free strains were not detected.C. trachomatis was detected in 8/660 (1.21%) swabs and semen samples by ompA gene Hemi Nested-PCR. The prevalence of C. trachomatis in the female group was 1.14% (5/439) and 1.38% (3/221) in males, with no statistically significant differences, p=0.8086, OR= 0.84 (0.17-4.45, CI 95%). The genotypes identified by sequencing analysis were D 12,5% (1/8), E 12,5% (1/8), G 25% (2/8), L1 50% (4/8). Genotype L1 was detected in 1 CS, 1 US and 2 US+S; genotypes E and G were detected in 1 CS each one, while genotype D was found in 1 US+S (Table 1).Infertility was the only symptom presented by C. trachomatis positive patients.The phylogenetic analysis confirmed that all the eight sequences clustered with C. trachomatis. The ompA sequences of the C. trachomatis strains were highly homologous and shared more than 98% similarity with each other. Blast searches revealed that ARG CA699 CT, ARG CA 042 CT, ARG CA 696 CT, ARG CA 043 CT ompA gene sequences showed high homology with L1 sequences. The CA 740 strain showed homology with other sequences of D genotypes, while ARG CA 534 CT revealed higher homology with E genotypes; ARG CA 006 CT and ARG CA 159 CT presented some similarities when compared to G genotypes. According to the phylogenetic tree, genotypes were subdivided into three distinct groups, genotypes B, D, E, L1 and L2 comprised one group; F and G a second group, and A, C, H, Ia, J, K, and L3 constituted a third group (Fig. 1).
|
4. Discussion
- In this study, we found that the detection of C. trachomatis cryptic plasmid (7.27%) was significantly higher (p<0.0001) than omp A C. trachomatis (1.21%) gene Hemi Nested PCR; these results were associated to the highest sensitivity of the cryptic plasmid PCR. It has been demonstrated that during the chlamydial developing cycle, up to 7.6 plasmids per chromosome can be detected, indicating an increased plasmid copy number in the actively replicating reticulate body [23]. The strong selection to maintain the plasmid by human chlamydial strains is related to its importance in the pathogenesis of human infections or diseases; however a fundamental ambiguity of C. trachomatis biology is the unknown function of cryptic 7.5-kb plasmid[5]. In addition, we did not detect cryptic plasmid free strains; similarly, Cuffini et al[7] found no such strain in a population of young asymptomatic people of Córdoba, Argentina. These results are important since the circulation of these variants is still unknown in most parts of the world.The prevalence of cryptic plasmid C. trachomatis was 7.52% in women and 6.79% in men. Our results are similar (percentage, studied population and methods) to a study performed in Poland, which reports a prevalence of 8.3% in women with infertility[27]. In American subjects, a study published last year in Mexico[9] described a prevalence of 15.8 % in women with infertility. Nevertheless, higher prevalence rates (43.3%) have been reported for asymptomatic male partners of infertile couples in Africa, Tunisia[13] and infertile women of Brazil (52.8%)[18]. The high prevalence may be explained because these patients had previous history of STI. In Córdoba, the prevalence of C trachomatis registered in asymptomatic sexually active young people and adolescents in 2008 was 8.7% (women: 13.7%, men: 4.1%) [10]. However, we detected a lower prevalence in our population of patients with infertility. This may be due to differences on the age range of the studied groups. Also, we lack information about the fertility status of the group of young people analyzed in the previous study.In this study, we also present the genotype distribution of C. trachomatis in these patients. Eight (8/600) C. trachomatis-positive samples were classified by phylogenetic analysis, demonstrating the presence of genotypes L1 (4/8), G (2/8), D (1/8) and E (1/8). The prevalence of C. trachomatis genotypes has been identified in several countries, with the genotype D (5-48%), D variants, E (22-44%) and F (8-20%) predominating in urogenital infections, while G (4-7%), Ga, H (<5%), I (6%), I variants, J (5-13%), and K (5-10%) were less common. Sporadically, genital infections with genotypes B and Ba also occur.[9, 19, 11, 15]. There are not many Latin-American studies of the distribution of C. trachomatis genotypes in infertile patients. A recent study in Mexico[9] pointed genotypes F (54.2%), E (8.7%), G (8.7%) and L2 (8.7%) as the most frequently found in women with infertility. In this study, we found genotypes E and D, and other genotypes less common, such as G and L1. Genotypes E and G were only found in women (12.5% and 25%, respectively) and genotype D was only detected in men (12.5%). L1 was found in both sexes but presented a higher proportion in men (12.5% vs. 37.5%). In Córdoba, a previous study in asymptomatic adolescents and young people pointed genotype E as the most common (57.14 of 73%) in women, followed by genotype D (16.2%); genotypes F and G were detected in lower proportion (5.4%) in both genders[7]. The same genotypes E, D and F were found in Buenos Aires city by Gallo Vaulet et al[11] in asymptomatic women, with a prevalence of 46.9%, 21% and 16.1%, respectively. We also detected circulation of genotype E in infertile patients, although in lower proportion than Cuffini et al[7]. We consider that this may be due to different characteristics of the studied groups (age, infertility). Surprisingly, we detected a high proportion (4/8) of L1 genotype in patients without symptoms of LGV, similarly to De Haro-Cruz et al, in Mexico[9], who also detected the genotype corresponding to LGV by phylogenetic analysis.Interestingly, the phylogenetic analysis showed three subdivisions, but the main branches did not coincide with the tissue tropisms and patterns of clinical presentation associated with C. trachomatis infection in human hosts. Identical results were obtained by Brunelle and Sensabaugh, and Lutter[2, 3, 19] in the phylogenetic characterization of the ompA gene. It has been proposed that the variability of MOMP is due to the antigenicity of MOMP and selective pressure of the immune system and as a result, the phylogeny of MOMP is not in agreement with tissue tropism or disease[19].C. trachomatis is a prevalent sexually transmitted infection that can lead to serious reproductive morbidity. The management and control of C. trachomatis is a challenge, largely due to its asymptomatic nature and our incomplete understanding of its natural history. Although chlamydia screening programs have been implemented worldwide, several countries have observed increasing rates of reported chlamydia new cases[26]. In conclusion, this is the first article from this region showing the prevalence rate of C. trachomatis infection in infertile patients and provides information on circulating genotypes. In addition, we demonstrated the absence of detection of cryptic plasmid free strains in this population in Cordoba; this constitutes an important data in the routine diagnosis. Local data also supports the need of a more extensive screening for infertility caused by C. trachomatis in Cordoba, with the intention of early detection, treatment and prevention.There are no conflicts of interests related to this study.
ACKNOWLEDGMENTS
- This study was supported in part by ad hoc research grants from A. Roemmers Foundation (2012-14).
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML