-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Probability and Statistics
p-ISSN: 2168-4871 e-ISSN: 2168-4863
2016; 5(3): 82-88
doi:10.5923/j.ijps.20160503.03

Generalized Linear Mixed Models for Longitudinal Data with Missing Values: A Monte Carlo EM Approach
Mohamed Y. Sabry , Rasha B. El Kholy , Ahmed M. Gad
Statistics Department, Faculty of Economics and Political Science, Cairo University, Cairo, Egypt
Correspondence to: Ahmed M. Gad , Statistics Department, Faculty of Economics and Political Science, Cairo University, Cairo, Egypt.
| Email: |  |
Copyright © 2016 Scientific & Academic Publishing. All Rights Reserved.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Longitudinal data have vast applications in medicine, epidemiology, agriculture and education. Longitudinal data analysis is usually characterized by its complexity due to inter-correlation between repeated measurements within each subject. One way to incorporate this inter-correlation is to extend the generalized linear model (GLM) to the generalized linear mixed model (GLMM) via including the random effects component. When the distribution of the random effects is far from the usual features of the Gaussian density, such as the student's t-distribution, this increases the complexity of the analysis as well as introducing critical features of subject heterogeneity. Thus, statistical techniques based on relaxing normality assumption for the random effects distribution are of interest. This paper aims to find the maximum likelihood estimates of the parameters of the GLMM when the normality assumption for the random effects is relaxed. Estimation is done in the presence of missing data. We assume a selection model for longitudinal data with a dropout pattern. The proposed estimation method is applied to both simulated and real data sets.
Keywords: Breast Cancer, Longitudinal data, Missing data, Mixed models, Monte Carlo EM
Cite this paper: Mohamed Y. Sabry , Rasha B. El Kholy , Ahmed M. Gad , Generalized Linear Mixed Models for Longitudinal Data with Missing Values: A Monte Carlo EM Approach, International Journal of Probability and Statistics , Vol. 5 No. 3, 2016, pp. 82-88. doi: 10.5923/j.ijps.20160503.03.
Article Outline
1. Introduction
- Longitudinal studies are not uncommon in biomedical research and social sciences. In such studies each individual (experimental unit) is measured repeatedly over time for the same response variable (outcome) either under different conditions, or at different times, or both. Longitudinal studies are subject to missing values. The missing values are termed dropout if the subject withdraws from the study at certain time without any observed value for it after that specified time. This is called monotone pattern for missing data. If the subject withdraws occasionally, then we have intermittent missing process and a non-monotone pattern.In the presence of missing data, the validity of any estimation method requires certain assumptions about the reasons behind the missingness; which are the missing data mechanism. Incorporating the missing data mechanism in statistical model means including an indicator variable, R, that takes the value 1 if an item is missing and 0 otherwise. Following Rubin's taxonomy [1], the missing data mechanism is said to be missing not at random (MNAR) if R depends on missing data and may depend on the observed data. In likelihood context this mechanism is labeled as non-ignorable. The likelihood estimation, especially in the presence of missing values, is intractable as integration over the random effects is required. Moreover, assuming non-normal distribution for the random effects makes the likelihood more complex and it cannot be expressed in closed form. Various approximation methods to evaluate the likelihood integrations were introduced. These include analytical approximation such as the penalized quasi-likelihood (PQL) and numerical integration such as Gauss-Hermite quadrature. Also a lot of iterative simulation techniques based on the EM algorithm have been used to optimize the likelihood function. The Monte Carlo EM algorithm was introduced to estimate likelihood parameters specially when sampling from conditional distribution of random effects or missing data is difficult. Violation of the normality assumption of random effects has been studied in literature. [2] uses a mixture of normals for the random effects density. [3] and [4] assume that the random effects distribution has a smooth but unspecified density, that can be estimated using semi-nonparametric representation. [5] replaces the normal distribution in linear mixed models by a skew-t distribution. [6] propose replacing the normal distribution with a mixture of normal distributions where the weights of the mixture components are estimated using a penalized approach. They also use the log-normal and gamma distribution for the random effects density. [7] estimate the maximum likelihood parameters for the logistic model when random effects follow a log normal distribution using a Monte Carlo EM (MCEM) algorithm. Many studies have been emerged focusing on estimating the GLMM parameters in the presence of missing data. [8] use a shared parameter model where an individual's random slope has been used as a covariate in a probit model for the censoring process. [9] propose conditioning on the time to censoring, and use censoring time as a covariate in random effects model. [10] approximate the generalized linear model using a generalized linear mixed model by conditioning the response variable on the missingness data. [11] develop a Monte Carlo EM algorithm to estimate parameters of GLMM with non-ignorable missing response data, with a normal random effects model. [12] propose a latent class model, which is an alternative approach to a pattern mixture model, by assuming that the dropout time belongs to unobserved (latent) class membership. [13] develop the stochastic EM-algorithm (SEM) algorithm for parameter estimation of a GLM in the presence of intermittent non-random missing values. [14] propose a pseudo likelihood method to estimate parameters for longitudinal data with binary outcome and categorical covariates. They assume missing not at random mechanism and non-monotone pattern. [15] propose a class of unbiased estimating equations using the pairwise conditional technique to estimate the GLMM parameters under non ignorable missingness with a normal random effects model. [16] propose a modification to the weights in the weighted generalized estimating equations to obtain unbiased estimates. [17] propose a heterogeneous random effects covariance matrix, which depends on covariates, using the modified Cholesky decomposition. [18] develop the hierarchical-likelihood method for nonlinear and generalized linear mixed models with arbitrary non-ignorable missing pattern and measurement errors in covariates.The maximum likelihood estimation of GLMM models in presence of missing data and non-normal distribution of the random effects is still a hot area of research. The aim of this paper is to propose a Monte Carlo EM algorithm to obtain the maximum likelihood estimates of GLMM model for longitudinal data. The focus is on longitudinal data with non-ignorable missingness. A selection model for longitudinal is used, where the responses are not normally distributed.The rest of the paper is organized as follows. Section 2 gives a description of the used model and notation. Section 3 is devoted to the novel estimation procedure. The performance of the proposed technique is evaluated via simulation studies in Section 4. In Section 5 the proposed technique is applied to a real data example. Finally, Section 6 presents the discussion.
2. Model and Notation
- The dependent outcome
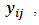
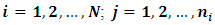 represent the jth measurement on the ith subject. Their distribution belongs to the exponential family and it can be expressed in a generalized linear model (GLM) as follows:
represent the jth measurement on the ith subject. Their distribution belongs to the exponential family and it can be expressed in a generalized linear model (GLM) as follows: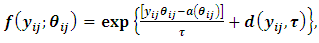 | (1) |
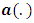 and
and 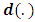 are functions defined according to the chosen distribution of exponential family and
are functions defined according to the chosen distribution of exponential family and 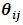 is called the natural parameter of the distribution. For simplification we assume a balanced design, that is
is called the natural parameter of the distribution. For simplification we assume a balanced design, that is 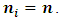 Let
Let 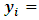
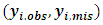 where
where 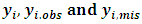 are complete, observed and missing outcomes vectors of subject i respectively. Assuming that we have
are complete, observed and missing outcomes vectors of subject i respectively. Assuming that we have 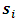 missing values for each subject i, then
missing values for each subject i, then 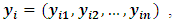
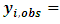
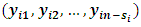 and
and 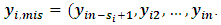 The conditional distribution of
The conditional distribution of 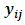 given the random effects
given the random effects 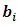 has a GLM form and follows the exponential family form as in Eq. (1) with a canonical identity link function, i.e.,
has a GLM form and follows the exponential family form as in Eq. (1) with a canonical identity link function, i.e., 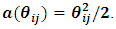 Thus
Thus 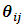 can be written in the form:
can be written in the form: 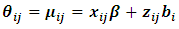 , where
, where 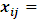
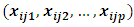 is the jth row of the
is the jth row of the 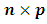 design matrix,
design matrix, 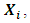 for vector of fixed effect parameters
for vector of fixed effect parameters 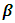 and
and 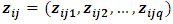 is the jth row in
is the jth row in 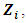 the design matrix of ith subject's random effect,
the design matrix of ith subject's random effect, 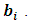 The scale parameter is
The scale parameter is 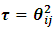 and
and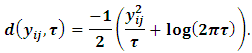 The GLMM can be written in the following form:
The GLMM can be written in the following form: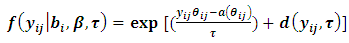 | (2) |
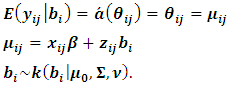 | (3) |
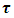 is constant and equals to one. Hence we can write
is constant and equals to one. Hence we can write 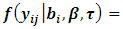
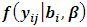 and
and 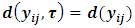 in Eq. (2). The function k(.) is the density function of the random effects
in Eq. (2). The function k(.) is the density function of the random effects 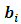 with non-central mean
with non-central mean 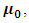 variance covariance matrix
variance covariance matrix 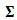 and
and 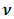 degrees of freedom.Note that the responses at all occasions j=1,...,n for any subject i are conditionally independent given the common random effect
degrees of freedom.Note that the responses at all occasions j=1,...,n for any subject i are conditionally independent given the common random effect 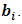 Thus, the likelihood for N subjects with complete data
Thus, the likelihood for N subjects with complete data 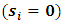 can be written as follows:
can be written as follows: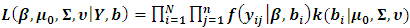 | (4) |
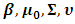 is proportional to the marginal density function
is proportional to the marginal density function 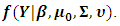 This can be obtained by integrating out the random effect, i.e.
This can be obtained by integrating out the random effect, i.e.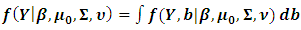 | (5) |
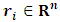 be the missing data mechanism indicator whose jth component
be the missing data mechanism indicator whose jth component 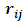 equals to 1 when the response
equals to 1 when the response 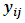 is missing, otherwise it equals to zero. Under the selection model [19], the distribution of
is missing, otherwise it equals to zero. Under the selection model [19], the distribution of 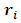 given the response
given the response 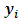 is indexed with a parameter vector
is indexed with a parameter vector 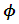 and has a multi-nominal distribution with
and has a multi-nominal distribution with 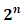 cell probabilities, i.e.
cell probabilities, i.e.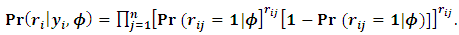 | (6) |
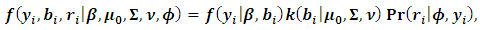 | (7) |
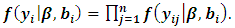 The classical normality assumption of the random effects is not always realistic. The t-distribution offers a more viable alternative with respect to real-world data. Assuming a central multivariate t-distribution
The classical normality assumption of the random effects is not always realistic. The t-distribution offers a more viable alternative with respect to real-world data. Assuming a central multivariate t-distribution 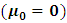 for the random effect,
for the random effect, 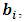 the density function k(.) is of the form
the density function k(.) is of the form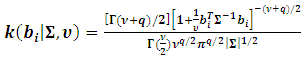 | (8) |
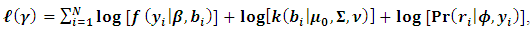 | (9) |
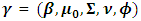 is the set of all parameters.
is the set of all parameters.3. Likelihood Estimation
- In this section, the fixed effect parameters,
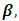 the random effects parameters,
the random effects parameters, 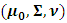 and the missing data model's parameters,
and the missing data model's parameters, 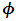 are estimated. A monotone pattern of missing data for the outcome variable
are estimated. A monotone pattern of missing data for the outcome variable 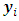 is assumed, where
is assumed, where 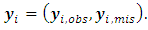 The random effects of the ith subject,
The random effects of the ith subject, 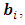 are unobserved, so they are also considered as missing data. Thus, in E-step of EM algorithm we need to integrate the complete-data log-likelihood in Eq. (9) over the missing data
are unobserved, so they are also considered as missing data. Thus, in E-step of EM algorithm we need to integrate the complete-data log-likelihood in Eq. (9) over the missing data 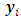 and
and 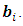 In other words, in the E step the expectation of the log-likelihood function of complete data given the observed data and current estimate of parameters is obtained. The expectation is with respect to the joint distribution of missing data
In other words, in the E step the expectation of the log-likelihood function of complete data given the observed data and current estimate of parameters is obtained. The expectation is with respect to the joint distribution of missing data 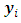 and
and 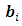 . Starting with initial values
. Starting with initial values 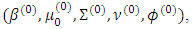 the (t+1)th iteration of the EM algorithm proceeds as follows:Ÿ The E-step: Obtain the expectations with respect to the conditional distribution of
the (t+1)th iteration of the EM algorithm proceeds as follows:Ÿ The E-step: Obtain the expectations with respect to the conditional distribution of 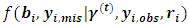 given in items 1 and 2 below. Also, obtain the conditional expectation with respect to conditional distribution of
given in items 1 and 2 below. Also, obtain the conditional expectation with respect to conditional distribution of 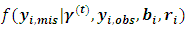 given in item 3 below.
given in item 3 below.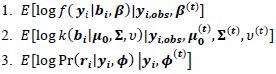 Ÿ The M step: Find the values
Ÿ The M step: Find the values 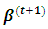 that maximize the function in item 1 above. Also, the values
that maximize the function in item 1 above. Also, the values 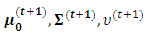 that maximize the function in item 2 above and
that maximize the function in item 2 above and 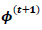 that maximize the function in item 3 above. If convergence is achieved, the current values are the maximum likelihood estimates. Otherwise, let
that maximize the function in item 3 above. If convergence is achieved, the current values are the maximum likelihood estimates. Otherwise, let 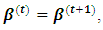
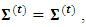
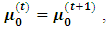
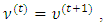
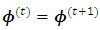 and return to the E-step.The expectation of the log-likelihood for the ith subject at iteration (t+1) is
and return to the E-step.The expectation of the log-likelihood for the ith subject at iteration (t+1) is 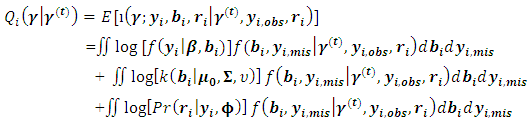 | (10) |
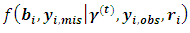 is the conditional distribution of missing data
is the conditional distribution of missing data 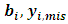 given the observed data and the current parameter estimates. The integral in Eq. (10) is intractable. Therefore, expectations are obtained via Monte Carlo simulation where sampling from
given the observed data and the current parameter estimates. The integral in Eq. (10) is intractable. Therefore, expectations are obtained via Monte Carlo simulation where sampling from 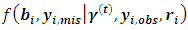 is required.The following two relations describe the complete conditional distributions
is required.The following two relations describe the complete conditional distributions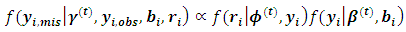 | (12) |
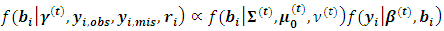 | (13) |
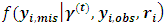 because the missingness mechanism doesn't depend on the random effects. A single draw
because the missingness mechanism doesn't depend on the random effects. A single draw 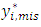 for the first dropout of the ith subject is generated from the predictive distribution
for the first dropout of the ith subject is generated from the predictive distribution 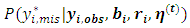 instead of
instead of 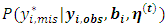 where
where 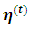 are the current parameter estimates. Then an accept-reject sampling procedure is used to sample
are the current parameter estimates. Then an accept-reject sampling procedure is used to sample 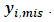 For more details, interested readers are referred to [20]. From properties of a multivariate normal,
For more details, interested readers are referred to [20]. From properties of a multivariate normal, 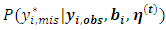 is a normal distribution with mean
is a normal distribution with mean 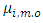 and covariance matrix
and covariance matrix 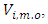 where
where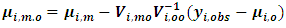 | (13) |
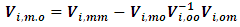 | (14) |
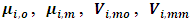 and
and 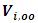 are suitable partitions from mean
are suitable partitions from mean 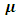 and covariance matrix V respectively. Sampling from the second complete conditional distribution in Eq. (12) is conducted using the Metropolis algorithm. A candidate value
and covariance matrix V respectively. Sampling from the second complete conditional distribution in Eq. (12) is conducted using the Metropolis algorithm. A candidate value 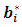 is generated from a proposal distribution which is chosen to be the density function of the random effects (the multivariate t). The generated value is accepted as opposed to keep the previous value with the probability:
is generated from a proposal distribution which is chosen to be the density function of the random effects (the multivariate t). The generated value is accepted as opposed to keep the previous value with the probability: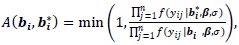 | (15) |
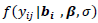 is a univariate normal and could be easily evaluated. The acceptance probability can be simplified as
is a univariate normal and could be easily evaluated. The acceptance probability can be simplified as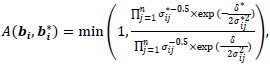 | (16) |
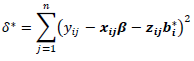 and
and 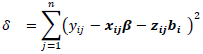 The
The 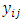 is jth row of
is jth row of 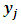 and
and 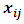 is the jth row in matrix
is the jth row in matrix 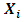 of dimension
of dimension 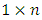 and
and 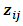 is the jth row in matrix
is the jth row in matrix 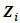 of dimension
of dimension 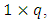
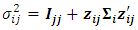 | (17) |
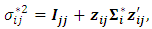 | (18) |
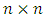 and
and  is the variance covariance matrix of random effects component
is the variance covariance matrix of random effects component 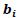 of dimension
of dimension 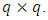
|
4. Simulation Study
- The aim of this simulation study is to evaluate the performance of the proposed techniques in Section (3). The number of subjects is assumed to be N = 30 subjects. The number of time points is assumed to be m = 7 time points. These numbers are chosen arbitrary for the sake of time. Hence, the response values are
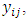 i = 1,2,…, 30, j=1,2,…,7. The random effects
i = 1,2,…, 30, j=1,2,…,7. The random effects 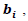 i = 1,2,…, 30, were generated from the multivariate t-distribution with dimension q =4 and known degrees of freedom
i = 1,2,…, 30, were generated from the multivariate t-distribution with dimension q =4 and known degrees of freedom 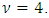 The response values were generated, conditionally on the random effect, with variance covariance matrix
The response values were generated, conditionally on the random effect, with variance covariance matrix 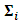 which is assumed to have a first order autoregressive correlation structure AR(1). The responses of the ith are modeled using the GLMM as
which is assumed to have a first order autoregressive correlation structure AR(1). The responses of the ith are modeled using the GLMM as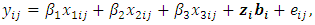 | (19) |
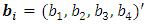 is a subject random effects. The
is a subject random effects. The 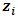 is a vector of ones
is a vector of ones 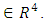 The noise component
The noise component 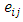 follows a multivariate normal
follows a multivariate normal 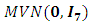 . The
. The 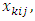 k=1,2,3; i=1,2,...,N; j=1,2,…,m are covariates (design matrix) associated with each subject. The values of
k=1,2,3; i=1,2,...,N; j=1,2,…,m are covariates (design matrix) associated with each subject. The values of 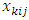 are chosen as follows:
are chosen as follows:  The missing data process is modeled as a logistic model, where the missing data mechanism depends on the current and previous outcome as:
The missing data process is modeled as a logistic model, where the missing data mechanism depends on the current and previous outcome as: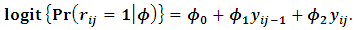 | (12) |
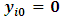 for all i. Also,
for all i. Also, 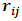 are assumed to be independent for all (i,j) given
are assumed to be independent for all (i,j) given 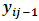 and
and 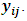 From Eq. (6) the missing data model for all observations can be written as:
From Eq. (6) the missing data model for all observations can be written as: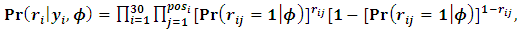 | (21) |
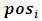 is the occasion at the first dropout for subject i.The parameter values are fixed as
is the occasion at the first dropout for subject i.The parameter values are fixed as 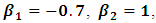
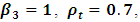
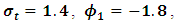
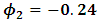 and
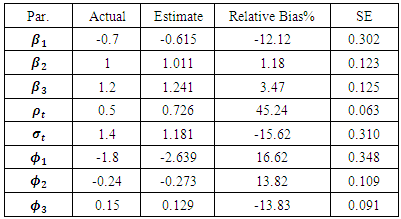
and 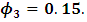 Table 1 shows the maximum likelihood estimates obtained using the MCEM algorithm, and the corresponding standard errors. 3000 samples were generated from missing data (missing outcome and random effect) with a burn-in period 500 iterations. Percentage of complete subjects is around 50%. As can be seen from Table 1, all parameters estimates are highly significant. All parameter estimates do not exceed a relative bias of 30% except
Table 1 shows the maximum likelihood estimates obtained using the MCEM algorithm, and the corresponding standard errors. 3000 samples were generated from missing data (missing outcome and random effect) with a burn-in period 500 iterations. Percentage of complete subjects is around 50%. As can be seen from Table 1, all parameters estimates are highly significant. All parameter estimates do not exceed a relative bias of 30% except 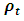 its bias percentage is 43.14%, where the relative bias is defined as the ratio of bias with respect to actual values. Small values for standard errors show that we have good results, which are lying in confidence interval 95%.All parameters of missing mechanism are significant, while parameter estimates of missing-mechanism for previous and current outcome are very significant, which support that the missing data-mechanism is non-ignorable.
its bias percentage is 43.14%, where the relative bias is defined as the ratio of bias with respect to actual values. Small values for standard errors show that we have good results, which are lying in confidence interval 95%.All parameters of missing mechanism are significant, while parameter estimates of missing-mechanism for previous and current outcome are very significant, which support that the missing data-mechanism is non-ignorable.5. Breast Cancer Data
- This data set is related to quality of life among premenopausal women diagnosed with breast cancer. It is prepared via a clinical trail taken by the International Breast Cancer Study Group (IBCSG) [21]. The patients are followed for death, relapse and quality of life. The patients were randomized to one of four chemotherapy regimes. The patients were asked to answer a quality of life questionnaire at baseline (before starting treatment) and every three months for fifteen months. Therefore, each questionnaire should be answered 6 times in the specified period.
|
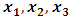 are defined as follows:
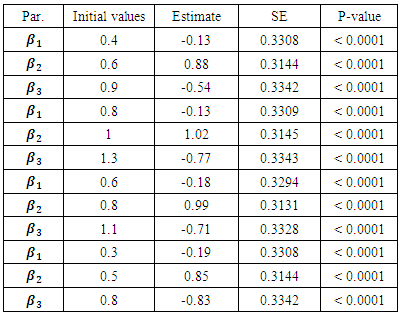
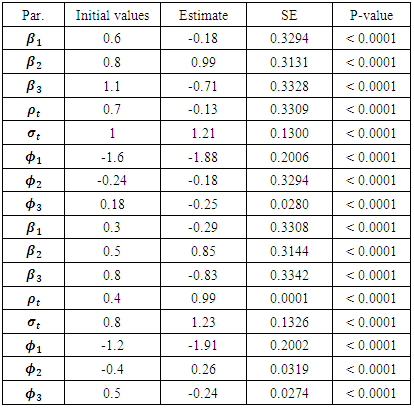
are defined as follows: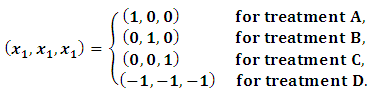 The MCEM is implemented using 3000 samples with a bur-in period of 500 iterations.
The MCEM is implemented using 3000 samples with a bur-in period of 500 iterations.
|
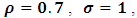
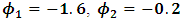 and
and 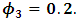 The estimates of
The estimates of 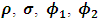 and
and 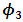 were around 0.99, 1.21, -1.90, 0.26, and -0.24 respectively. The results in Table 3 are the maximum likelihood estimates using different starting points for all parameters. Results show that the algorithm converges properly as the parameter estimates are similar for the different starting values. Also, all estimates are highly significant. Highly significant coefficients of treatments are indication of strong relationship between the response variable (PACIS) and the treatments. Given, that any patient receives only one treatment during the trail, the higher doze from treatment A or D, the smaller PACIS value i.e. a better quality of life the patient gets. On the other hand, the higher doze from treatment B or C, the higher PACIS value, i.e. patient endure hardship to cope with her life. Also, the significance of
were around 0.99, 1.21, -1.90, 0.26, and -0.24 respectively. The results in Table 3 are the maximum likelihood estimates using different starting points for all parameters. Results show that the algorithm converges properly as the parameter estimates are similar for the different starting values. Also, all estimates are highly significant. Highly significant coefficients of treatments are indication of strong relationship between the response variable (PACIS) and the treatments. Given, that any patient receives only one treatment during the trail, the higher doze from treatment A or D, the smaller PACIS value i.e. a better quality of life the patient gets. On the other hand, the higher doze from treatment B or C, the higher PACIS value, i.e. patient endure hardship to cope with her life. Also, the significance of 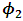 and
and 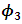 supports that the missing data mechanism is non-ignorable.
supports that the missing data mechanism is non-ignorable.6. Discussion
- In this paper, a Monte Carlo EM algorithm is proposed to estimate the likelihood parameters of a longitudinal model in GLMM framework. We used a selection model for longitudinal data with non-ignorable missing mechanism in the context of dropout pattern. In this case the obtained likelihood function is intractable and it is difficult to integrate over the missing data (random effect and missing values of outcomes). Therefore a Monte Carlo EM algorithm is implemented. In the context of the proposed model, direct simulation is not possible because there is no defined mathematical form for the joint density of missing data (random effect and missing outcomes) given the observed data and current estimate of parameters. Thus, Monte Carlo Sampling techniques are used; Metropolis-Hastings and reject-accept techniques. Hessian matrix is used to estimate standard errors. The proposed algorithm is validated using a simulation study. Also, the proposed algorithm is applied to a real data set of breast cancer.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML