-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Modern Botany
p-ISSN: 2166-5206 e-ISSN: 2166-5214
2016; 6(2): 31-36
doi:10.5923/j.ijmb.20160602.03

Simple and Quantitative Detection of Apple latent spherical virus Vector by a Spot Hybridization
Sachin Ashok Bhor1, Md. Shamim Akhter1, Takashi Yaeno1, Naoto Yamaoka1, Masamichi Nishiguchi2, Masanori Kaido3, Kappei Kobayashi1
1The United Graduate School of Agricultural Sciences, Ehime University, Matsuyama, Ehime, Japan
2Faculty of Agriculture, Ehime University, Matsuyama, Ehime, Japan
3Laboratory of Plant Pathology, Graduate School of Agriculture, Kyoto University, Kyoto, Japan
Correspondence to: Kappei Kobayashi, The United Graduate School of Agricultural Sciences, Ehime University, Matsuyama, Ehime, Japan.
| Email: |  |
Copyright © 2016 Scientific & Academic Publishing. All Rights Reserved.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Apple latent spherical virus (ALSV) vector has been widely used as a vector for virus-induced gene silencing, gene expression, and vaccination for protection from virus diseases in wide variety of plants. Here we describe a simple and quantitative method for ALSV detection in plant materials. ALSV infected plant leaves were homogenized in a buffer, and added with NaOH, which was found to be optimal at the final concentration of 10 mM. After serial dilution with 10 mM NaOH, the samples were spotted onto nylon membrane. After neutralization with acidic solution and fixation of nucleic acid by UV irradiation, the membrane was subjected to a prehybridization for an hour, followed by a hybridization with digoxygenin-labelled RNA probe for overnight. ALSV was quantitatively detected using a chemiluminescent substrate. The entire protocol took two days with hands-on time of a couple of hours.
Keywords: ALSV, Hybridization, Non-Radioactive probe, VIGS
Cite this paper: Sachin Ashok Bhor, Md. Shamim Akhter, Takashi Yaeno, Naoto Yamaoka, Masamichi Nishiguchi, Masanori Kaido, Kappei Kobayashi, Simple and Quantitative Detection of Apple latent spherical virus Vector by a Spot Hybridization, International Journal of Modern Botany, Vol. 6 No. 2, 2016, pp. 31-36. doi: 10.5923/j.ijmb.20160602.03.
Article Outline
1. Introduction
- Plant virus vectors have been widely used for the functional analysis of plant genes with a great advantage over the production of stably transformed plants. Wild-type plants grown from seeds can be infected with recombinant viruses harbouring a foreign gene or a gene from the same plant to overexpress the protein product, which takes much lesser time than the genetic transformation. Virus-induced gene silencing (VIGS) is a widely used method for loss-of-function analysis in plant biology, which takes much lesser time than the establishment of stable transformants expressing antisense or hairpin RNAs. Studies have utilized high-levels expression from virus vectors for identifying and analysing plant genes involved in cell death [1-3], and others have identified genes involved in disease resistance or susceptibility using VIGS [4, 5]. Different plant viruses have been manipulated to construct virus vectors [6]. Among them, Apple latent spherical virus (ALSV) vector has a great advantage over other virus vectors in its broad host range [7-11]. Moreover, ALSV vector has been shown to be useful for vaccination of plants to prevent and treat different virus diseases [12-14].The infection of plants with ALSV can efficiently be achieved by a mechanical inoculation of plasmid DNA or agroinfection in many plant species, but is less efficient in some plant species. Alternative inoculation methods include the particle bombardment of RNA from ALSV-infected plants [8-11], and the mechanical inoculation of crude homogenate of ALSV-infected plants [7]. To infect plants with ALSV in these methods in a highly reproducible manner, it is of great importance to measure the virus titre prior to the inoculation of plants with RNA or crude homogenate. To this end, we applied a spot hybridization [15]. Here we report a simple spot hybridization technique optimized for ALSV quantification.
2. Methods
2.1. Plant Growth
- Nicotiana benthamiana and Nicotiana tabacum seeds were sown onto two layers of wetted filter paper and kept at 25°C with 16 hrs light and 8 hrs dark periods. One week later, the seedlings were transferred to Jiffy-7 (4.2 cm in diameter before swelling; Jiffy Products International, Kristiansand, Norway). The plants were grown at 25°C with 16 hrs light and 8 hrs dark periods for another 7—10 days before being inoculated with ALSV.
2.2. ALSV Vectors
- The binary vector-based plasmids, pBICAL1 and pBICAL2, which contain the expression units for cDNAs of ALSV RNA1 and RNA2, respectively, were described previously [9] (Fig. 1). For the silencing of tobacco defence-related genes, several pBICAL2-derivatives were constructed essentially as described previously [9]. Total RNA was extracted from tobacco (Nicotiana tabacum cv. SR1) leaf tissue using Sepasol-RNA 1 Super G (Nacalai tesque, Kyoto, Japan) as described previously [16]. A cDNA was synthesized using random hexamer primers and PrimeScript reverse transcriptase (Takara Bio, Ohtsu, Japan). Different gene fragments were amplified using the cDNA as templates, primers shown in Table 1, and PrimeSTAR GXL DNA polymerase (Takara Bio). The amplified fragments were cloned using Zero Blunt TOPO PCR cloning kit, sequenced, and excised by XhoI and BamHI digestion to be introduced into pBICAL2. The resulting plasmids were introduced into Agrobacterium tumefaciens GV3101.
|
2.3. Inoculation of Plants with ALSV
- A. tumefaciens harbouring pBICAL1, and pBICAL2 or its derivatives were propagated on an agar plate medium containing appropriate antibiotics, scraped off the plates using a plastic inoculation loop, and suspended in 10 mM K-MES, pH 5.5 containing 10 mM MgCl2. About 20 days old N. benthamiana seedlings were inoculated with mixtures of A. tumefaciens harbouring pBICAL1, and pBICAL2 or its derivatives using a toothpick. After one day at 22°C in 100% humidity, plants were returned to aforementioned growth condition and grown for another four weeks. In all experiments, ALSV-PDS, which derives from pBICAL1 and pBICAL2-PDS and induces photo-bleaching phenotype, was included as a positive control for VIGS.
2.4. Sample Preparation
- N. benthamiana leaves were harvested 3 weeks post-infection with ALSV vectors, and homogenized in two volume of extraction buffer (0.1 M Tris–HCl, pH7.8, 0.1 M NaCl, 5 mM MgCl2) in a sterilized mortar and pestle with sea sand C (Nacalai tesque). After cleared by centrifugation at 13,500 ×g for 10 min at 4°C, a small amount of crude sap was mixed with NaOH solution to final concentration of 10, or in some experiments 50 mM. The samples were serially two-fold or three-fold diluted with the same concentration of NaOH solution.
2.5. Membrane Preparation
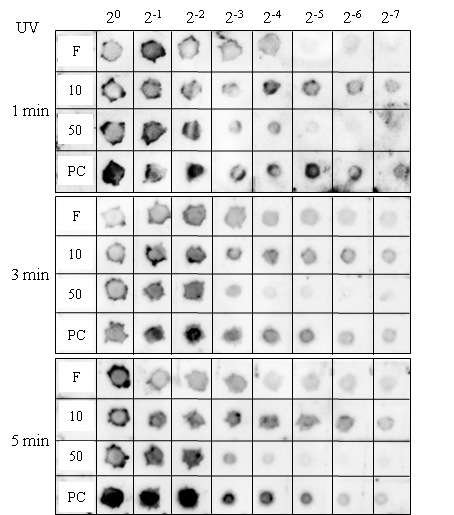
- A positively charged nylon membranes (Hybond N+, GE Healthcare) was cut at an appropriate size (e.g., 10 cm × 4 cm) and drawn with 1cm lattice with a pencil. The membrane was equilibrated first with 10× SSC and then with 20× SSC by placing on pieces of Whatman 3MM filter paper immersed with the solutions for 5 min each. After air-dried for 1 hr or more, the membrane was spotted with 2 µl of serially diluted samples. After spotting all the samples the membrane was air-dried for 2 hrs, and then exposed to UV light on a transilluminator for 1, 3 or 5 min to cross-link the nucleic acids to the membrane.
2.6. Probe Preparation
- The 841 bp fragment of ALSV-RNA2 was PCR amplified by using ALSV-R2-1679F (5´-TACCAGGGAGGAC CTTGTTG-3´) and T7-ALSV-R2-2520R (5´-GGTTAATA CGACTCACTATAGGGAAGCGAGTGCGAGAATAGG A-3´) primers and pBICAL2 plasmid (1 ng/µl) as a template. Digoxigenin (DIG)-labelled RNA probe was synthesized using DIG Northern Starter Kit (Roche). After the in vitro transcription at 37°C for 3 hrs, the template DNA was removed from the mixture by RNase-free DNase I treatment at 37°C for 15 min. The reaction was stopped by adding 2 µl, 0.2 M EDTA (pH 8.0).
2.7. Prehybridization, Hybridization and Detection
- The cross-linked membrane (10 cm × 4 cm) was sealed in a plastic bag with 10 ml of DIG Easy Hyb buffer (Roche) and incubated for 1 hr at 68°C. DIG-labelled RNA probe was denatured by boiling for 5 min and quickly chilling in ice water. Two microliter of denatured probe was mixed with 5 ml of prewarmed DIG Easy Hyb buffer, and used for hybridization for overnight at 68°C. The membrane was washed twice in 2× SSC containing 0.1% SDS with continuous shaking for 5 min at room temperature, and then twice for 15 min each in 0.1× SSC containing 0.1% SDS at 68°C. ALSV RNA was detected according to the manual for DIG Northern Starter Kit (Roche) with the CDP-Star chemiluminescent substrate. The chemiluminescent image was obtained using ChemiDocTM MP Imaging system (Bio-Rad).
3. Results and Discussion
3.1. Optimal Condition for ALSV Vector Detection by Spot Hybridization
- To optimize the detection procedure of spot hybridization for the ALSV vector quantification, we first used the ALSV-PDS, which was expected to induce visible photo-bleaching symptoms and therefore facilitates the evaluation of VIGS efficiency.3.1.1. ALSV-mediated VIGS of PDS Gene in N. benthamiana and N. tabacumThe ALSV-PDS that we constructed based on the PDS gene sequence of N. tabacum reproducibly induced the silencing of PDS gene expression and the visible photo-bleaching in both N. benthamiana and N. tabacum (Fig. 2).
3.2. Detection of ALSV Vector with Different Target Gene Inserts by Spot Hybridization
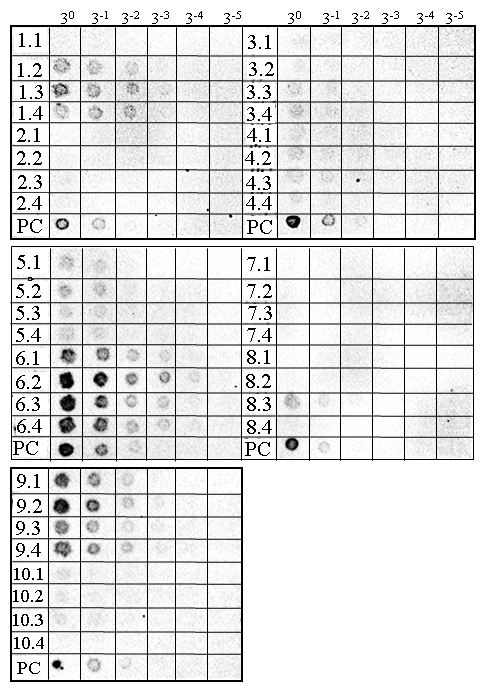
- Next we compared the accumulation levels of recombinant ALSV vectors harbouring different target gene inserts using the optimized technique. We observed high-level accumulation of recombinant ALSV only for those harbouring PDS or NDR1 gene segments in N. benthamiana plants (Fig. 4, PC and 6.1–6.4, respectively). Recombinant ALSVs harbouring either of ADR1, EDS1, NPR1, or NGR1 gene fragments were shown to accumulate to low levels in N. benthamiana plants (Fig. 4, 1.1–4, 3.1–4, 4.1–4, and 5.1–4). These results suggest that ALSV accumulation levels may vary among those harbouring different gene segments. Such reduction in the virus accumulation levels could be attributed to the silencing of the target genes, which might have roles in virus replication. In case of ADR1, we constructed recombinant ALSV with two different target regions, ADR1-1 and ADR1-2 (Table 1). We detected low levels of ALSV accumulation in three out of four crude virus preparations in ADR1-1, but failed to detect any sign of virus accumulation for ADR1-2, suggesting that the reduced ALSV accumulation should be attributed to the nature of inserted fragment rather than to the silencing of the target gene. This would be also the case for recombinant ALSV harbouring Rar1 gene segment (Fig. 4, 7.1-4), which we failed to detect any sign of virus accumulation. By contrast, we detected low-level accumulation of recombinant ALSV harbouring SGT1 gene segment (Fig. 4, 8.1-4), albeit in only one preparation out of four. This result suggests that some gene segments introduced into ALSV could affect virus replication in a stochastic manner.
4. Conclusions
- In this study, we optimized the quantitative detection of recombinant ALSV. This method would help us optimize the inoculation procedure to establish reliable VIGS assay using ALSV vectors.
ACKNOWLEDGEMENTS
- The authors thank N. Yoshikawa for generous gift of original ALSV vector clones. This study was supported by in part by United Graduate School of Agricultural Sciences, Ehime University, and JSPS KAKENHI grant No. 24658044 to K.K. SAB has been supported by Rotary Yoneyama Memorial Foundation, and MSA by MEXT, Japan.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML