-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Genetic Engineering
p-ISSN: 2167-7239 e-ISSN: 2167-7220
2026; 14(1): 7-12
doi:10.5923/j.ijge.20261401.02
Received: Sep. 28, 2025; Accepted: Oct. 22, 2025; Published: Jan. 9, 2026

Molecular Genetic Characterization of the Species Vespa crabro and Vespa orientalis Belonging to the Genus Vespa
Yuldosheva Jamila1, Bobonazarov Gappar1, Burieva Khurshida2, Amirov Oybek3, Shapaotov Ruziboy3
1Department of Zoology, Karshi State University, Karshi, Uzbekistan
2Department of Botany, Turon University, Karshi, Uzbekistan
3Laboratory of Molecular Zoology, Institute of Zoology, Tashkent, Uzbekistan
Correspondence to: Yuldosheva Jamila, Department of Zoology, Karshi State University, Karshi, Uzbekistan.
| Email: |  |
Copyright © 2026 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

This study presents a molecular-genetic characterization of two hornet species, Vespa crabro and Vespa orientalis, collected from the Qashqadaryo Region of Uzbekistan. Specimens were identified morphologically and analyzed using mitochondrial cytochrome oxidase I (COI) gene sequences to assess genetic differentiation within the genus Vespa. A 623 bp fragment of the COI gene was successfully amplified and sequenced from both species and compared with reference sequences available in GenBank (V. crabro MT444756; V. orientalis KF933083). The results revealed low levels of intraspecific variation, with Uzbek V. crabro and V. orientalis differing from their respective reference sequences by 0.2% and 0.3%, respectively. In contrast, interspecific comparisons showed clear genetic divergence, with COI sequence differences ranging from 2.7% to 3.04%. These values fall within the accepted thresholds for species-level discrimination in Vespidae and demonstrate consistent nucleotide polymorphisms distinguishing the two species. The findings confirm the effectiveness of COI barcoding for reliable identification and differentiation of V. crabro and V. orientalis and provide the first molecular genetic data for these species from Uzbekistan. This study contributes to filling a geographic gap in the molecular taxonomy of the genus Vespa and establishes a baseline for future phylogeographic and biodiversity studies in Central Asia.
Keywords: Vespa crabro, Vespa orientalis, Mitochondrial COI, Nucleotide sequence, Uzbekistan
Cite this paper: Yuldosheva Jamila, Bobonazarov Gappar, Burieva Khurshida, Amirov Oybek, Shapaotov Ruziboy, Molecular Genetic Characterization of the Species Vespa crabro and Vespa orientalis Belonging to the Genus Vespa, International Journal of Genetic Engineering, Vol. 14 No. 1, 2026, pp. 7-12. doi: 10.5923/j.ijge.20261401.02.
Article Outline
1. Introduction
- The European hornet, Vespa crabro, is the largest social wasp in Europe, with workers measuring up to 23 mm and queens up to 30 mm [1]. It is widespread across the Palearctic region, including England and Wales, and has been introduced into North America [2]. Vespa crabro occupies diverse habitats, but it is particularly associated with mature deciduous woodlands and urban environments. Morphologically, the head and most of the metasoma are predominantly yellow, whereas the mesosoma, anterior metasomal segments, and legs are reddish-brown with variable black markings.This species is eusocial, forming colonies with a queen, workers, and males [3]. Colony founding typically begins in early spring, and nests are constructed from a paper-like material produced by mixing decayed wood pulp with saliva [4,5]. Nests may contain multiple combs enclosed by an envelope and are usually located in hollow trees and, frequently, in human structures such as attics and outbuildings; nest relocation may occur when space becomes limiting [6,7]. Female hornets are generalist predators that capture a broad range of arthropod prey, including other social wasps, honeybees, flies, lepidopterans, and spiders [7-10]. They may also feed on plant exudates at sap runs and visit flowering plants for nectar [4,7,11]. Where V. crabro co-occurs with the invasive Vespa velutina, resource overlap may lead to interspecific competition [12]. Although generally not highly aggressive, stings can occur during nest defense, and comparative venomics indicates marked enrichment of major venom components in V. crabro relative to some congeners [13].The Oriental hornet, Vespa orientalis, is distributed across the Levant, Southern Europe, Northeast Africa, and Southwestern Asia, including India [14,20]. It is considered an important apicultural pest due to its strong predation pressure on honeybees and its potential to reduce foraging activity, leading to economic losses for beekeepers and decreased crop pollination [14]. In addition, it may damage fruit crops directly or indirectly, and stings can cause medically relevant reactions in sensitive individuals [14-16]. Several subspecies and regional forms have been described, often distinguished by coloration traits [17]. Previous studies have reported morphological features, including the sting apparatus and antennal chemoreceptors [18,19], and the complete mitochondrial genome of V. orientalis has been characterized [20].Despite the broad recognition and distribution of V. crabro [2], the taxonomic history of hornets has been complex, and subspecies delineation based on morphology-especially coloration-has been debated [21,22]. Recent taxonomic reassessments have questioned the reliability of some morphological characters and suggested that several subspecies names may represent synonyms, emphasizing the need for molecular evidence in species delimitation [23]. In this context, mitochondrial COI barcoding remains a widely applied and reliable approach for insect identification and comparative taxonomy.Therefore, the aim of this research is to provide a combined morphological and molecular-genetic characterization of Vespa crabro and Vespa orientalis from Uzbekistan using the mitochondrial COI gene, and to assess genetic differentiation between these two species within the genus Vespa [23].
1.1. Zoological Description
- Vespa crabroVespa crabro is one of the largest hornet species recorded in Uzbekistan. The body length of workers ranges from 18 to 24 mm, females measure 22-25 mm, and males 21-28 mm. The head is predominantly yellow, with the occipital and cervical regions showing a reddish hue in females, while males exhibit brighter yellow coloration. Mandibles are yellow, and antennae are reddish in females and yellow in males. The mesosoma is black with reddish markings, particularly on the pronotum. The metasoma is light yellow with distinct black spots, and the base of the first tergite is characteristically red. Legs are reddish, with the basal portions of the femora darkened.Sexual dimorphism is evident in antennal segmentation and abdominal structure: females possess 12 antennal segments and seven visible abdominal segments, whereas males have 13 antennal segments and six abdominal segments. Diagnostic characters of V. crabro include its large body size, contrasting yellow-red coloration, and robust body proportions. The species is a generalist predator, feeding on a wide range of insects, including bees and wasps [24,25].Vespa orientalisVespa orientalis is a large social wasp exhibiting considerable size variation, with total body length ranging from 19 to 32 mm. Females are the largest caste (25-32 mm), followed by males (20-25 mm) and workers (19-25 mm). The metasoma shows a characteristic coloration pattern, ranging from reddish-brown to dark brown, with the third (T3) and fourth (T4) tergites predominantly yellow and marked by a basal reddish-brown band extending medially, accompanied by two lateral spots. This color pattern represents a key diagnostic feature of the species.The posterior ocelli are positioned closer to each other than to the compound eyes. The clypeus lacks a medial tooth between the broadly rounded lateral projections, and the genae are as wide as or slightly wider (up to 1.5 times) than the compound eye in lateral view. A complete pretegular carina is present [26]. Sexual dimorphism is reflected in antennal segmentation, with males bearing 13 segments and females 12. The thorax is uniformly brown, and the wings measure 34-46 mm in length. The abdomen consists of six to seven visible segments, with males typically exhibiting seven segments.Geometric morphometric studies have demonstrated notable variation in forewing shape within V. orientalis, leading to the recognition of three geographic morphogroups: AFRI (North Africa), MEDI (Northern Mediterranean), and MEAS (Middle East). The AFRI morphogroup shows pronounced wing-shape divergence, likely influenced by geographic isolation and environmental adaptation, whereas MEDI and MEAS groups exhibit more moderate differentiation corresponding to regional boundaries [27]. These morphometric differences may influence dispersal and foraging behavior but do not obscure the core diagnostic morphological traits of the species.
2. Materials and Methods
- Entomological specimens were collected between 2022 and 2025 in the Kitab, Nishon, and Mirishkor districts of the Qashqadaryo Region, Uzbekistan. Adult individuals of Vespa crabro and Vespa orientalis (genus Vespa) were collected and preserved in 70% ethanol until further analysis.Genomic DNA was extracted from ethanol-preserved specimens using the GeneJET Genomic DNA Purification Kit (Germany), following the manufacturer’s protocol. The isolated genomic DNA was used to amplify a fragment of the mitochondrial cytochrome oxidase I (COI) gene, which is widely applied in insect molecular taxonomy. Polymerase chain reaction (PCR) amplification was performed using primers C1-J-2195(TTGATTTTTTGGTCATCCAGAAGT) and L2-N-3014 (TCCAATGCACTAATCTGCCATATTA), previously validated for insect COI barcoding (Frohlich et al., 1999).PCR reactions were carried out in a programmable thermal cycler (Touchgene Gradient, UK) under the following conditions: initial denaturation at 95°C for 3 min; followed by 35 cycles of denaturation at 93°C for 20 s, primer annealing at 55°C for 30 s, and elongation at 72°C for 2 min; with a final extension step at 72°C for 10 min. PCR products were visualized and purified, and DNA concentrations were measured using a NanoDrop spectrophotometer prior to sequencing.Sequencing was performed using the ABI PRISM® BigDye™ Terminator v.3.1 kit, and reaction products were analyzed on an ABI PRISM 3100-Avant automated sequencer (Core Facility “Genome”, Genotech, Moscow). Sequencing chromatograms were obtained in AB1 format and initially inspected using Chromas version 1.45 [28]. Forward and reverse sequences were assembled and converted to FASTA format, after which multiple sequence alignment was conducted using Clustal X version 1.81 [29]. Ambiguous or low-quality regions were removed using GeneDoc version 2.5.000 [30]. Sequence data were converted to Nexus format using ForCon version 1.0 for Windows.Phylogenetic and comparative analyses were performed using MEGA11 and PAUP* version 4.0b10 [31]. All PAUP* analyses were conducted using input files in Nexus format.
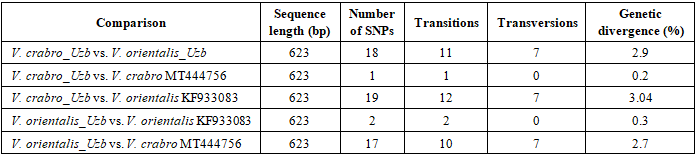
3. Results
- Based on molecular-genetic analyses of mitochondrial DNA, a 623 base-pair fragment of the cytochrome oxidase I (COI) gene was successfully amplified and sequenced from Vespa crabro and Vespa orientalis specimens collected in Uzbekistan. The obtained sequences were designated as V. crabro_Uzb and V. orientalis_Uzb. These sequences were compared with corresponding reference COI sequences available in GenBank, including V. crabro (accession number MT444756) and V. orientalis (accession number KF933083).Comparative analysis revealed clear nucleotide polymorphisms both within and between species. A total of 18 nucleotide substitutions were detected between V. crabro_Uzb and V. orientalis_Uzb across the analyzed 623 bp COI fragment. These substitutions included both transitions and transversions, with transitions occurring more frequently, which is consistent with typical mitochondrial DNA substitution patterns.Intraspecific comparison showed minimal genetic variation. The V. crabro_Uzb sequence differed from the reference V. crabro MT444756 by a single nucleotide substitution (0.2%), indicating high genetic similarity within the species. Similarly, V. orientalis_Uzb exhibited low divergence from the reference V. orientalis KF933083 sequence, differing by only two nucleotide substitutions (0.3%).In contrast, interspecific comparisons demonstrated substantially higher levels of divergence. The genetic distance between V. crabro_Uzb and V. orientalis reference sequences reached 3.04%, while V. orientalis_Uzb differed from V. crabro MT444756 by 2.7%. A summary of the number and type of nucleotide substitutions, as well as uncorrected genetic distances for all pairwise comparisons, is presented in Table 1.
|
4. Discussion
- The molecular-genetic analysis of the mitochondrial cytochrome oxidase I (COI) gene fragment from Vespa crabro and Vespa orientalis specimens collected in the Qashqadaryo Region of Uzbekistan revealed clear patterns of nucleotide polymorphism that contribute to understanding genetic differentiation within the genus Vespa. Sequencing of a 623 bp COI fragment and comparison with reference sequences from GenBank (V. crabro MT444756; V. orientalis KF933083) demonstrated distinct interspecific divergence alongside minimal intraspecific variation.A total of 18 nucleotide substitutions were detected between the local V. crabro_Uzb and V. orientalis_Uzb sequences, resulting in an uncorrected genetic distance of approximately 2.9%. This level of divergence falls within the range commonly reported for interspecific differentiation in Vespidae based on COI barcoding and is consistent with previous molecular studies of Vespa species. In contrast, intraspecific comparisons showed very low levels of variation: V. crabro_Uzb differed from the reference MT444756 sequence by a single nucleotide substitution (0.2%), and V. orientalis_Uzb differed from KF933083 by only two substitutions (0.3%). These findings indicate genetic stability within species and support the reliability of COI as a species-level diagnostic marker.The observed substitutions included both transitions and transversions, with transitions predominating, a pattern characteristic of mitochondrial DNA evolution. Several fixed nucleotide differences between V. crabro and V. orientalis were consistently detected across comparisons, providing clear molecular signatures for species discrimination. The multiple sequence alignment (Figure 1) highlights these species-specific single nucleotide polymorphisms while demonstrating limited variation within each species, reinforcing the conclusions drawn from pairwise distance analyses (Table 1).
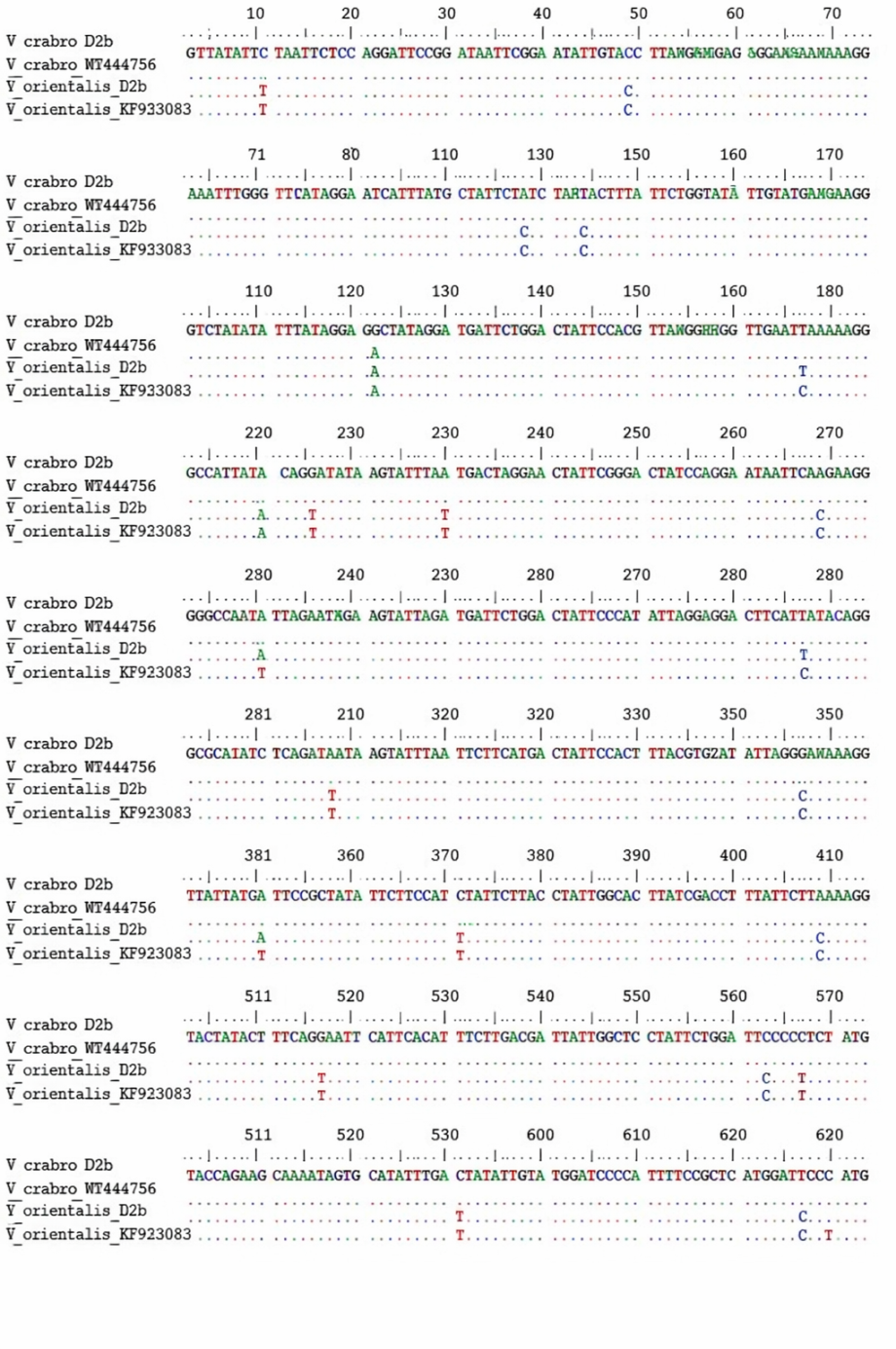 | Figure 1. Comparison of nucleotide substitutions in the 623 bp COI mitochondrial DNA fragment of Vespa crabro and Vespa orientalis, including Uzbek specimens and reference sequences from GenBank |
5. Conclusions
- This study provides the first molecular-genetic characterization of Vespa crabro and Vespa orientalis from Uzbekistan based on a 623 bp fragment of the mitochondrial cytochrome oxidase I (COI) gene. Comparative analysis with reference sequences from GenBank (V. crabro MT444756; V. orientalis KF933083) revealed very low levels of intraspecific variation (0.2-0.3%), indicating genetic stability within each species. In contrast, interspecific comparisons showed clear genetic differentiation, with uncorrected COI divergences ranging from 2.7% to 3.04%. These values fall within the commonly accepted thresholds for species-level separation in Vespidae and confirm the distinct genetic identities of V. crabro and V. orientalis. The observed nucleotide polymorphisms support the effectiveness of COI barcoding as a reliable tool for species discrimination within the genus Vespa.Overall, the results contribute new molecular data for Central Asian populations of hornets and help fill a geographic gap in the molecular taxonomy of Vespa. These findings provide a baseline for future studies addressing population structure, phylogeography, and the ecological and economic significance of hornets in Uzbekistan and adjacent regions.
ACKNOWLEDGEMENTS
- The author gratefully acknowledges the staff of the Molecular Zoology Laboratory, Institute of Zoology, Academy of Sciences of the Republic of Uzbekistan, for their technical assistance and support during the molecular genetic analyses of Vespa species.
DISCLOSURE
- The authors declare no conflicts of interest related to the research, authorship, or publication of this study. No external funding was received. All research was conducted in accordance with applicable ethical standards, and entomological specimens were collected with the necessary permits from relevant authorities in Uzbekistan. All nucleotide sequence data referenced in this study are publicly available in international databases, including GenBank (accession numbers MT444756 and KF933083).
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML