-
Paper Information
- Next Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Genetic Engineering
p-ISSN: 2167-7239 e-ISSN: 2167-7220
2025; 13(6): 130-132
doi:10.5923/j.ijge.20251306.09
Received: Jun. 2, 2025; Accepted: Jul. 6, 2025; Published: Jul. 11, 2025

Genetic Identification of Isolates of the Fungus Schizophyllum commune Fr. and Molecular Phylogenegy within the Species
Erkin Eshonkulov Yulchi ugli
Lecturer, Karshi State University, Karshi, Uzbekistan
Correspondence to: Erkin Eshonkulov Yulchi ugli, Lecturer, Karshi State University, Karshi, Uzbekistan.
| Email: |  |
Copyright © 2025 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

This article presents information on the identification of the 1,5.8S ribosomal RNA gene fragment from strains grown in liquid nutrient medium, the placement of nucleotide sequences in the gene bank, and the molecular phylogeny within the species of isolates isolated from the medicinal fungus Schizophyllum commune, which is distributed in Uzbekistan for the first time. The latest modern biotechnological, mycological, microbiological, and molecular genetic methods were used in the research work. As a result of the research, the fungal isolates were registered in the world-famous international gene banks, including NCBI, the European Nucleotide Archive, and the DDBJ Japanese DNA database with the ID numbers JE4 strain PP837386 and JE5 strain PP837387. These analyses allowed us to determine the specificity of S.commune fungal strains and, based on the nucleotide sequences of these strains, to construct an intraspecific molecular phylogenetic tree of strains occurring in different regions of the world and to compare isolates.
Keywords: Fungus, Isolate, Phylogenetic tree, Supernatant, Gene, Fragment
Cite this paper: Erkin Eshonkulov Yulchi ugli, Genetic Identification of Isolates of the Fungus Schizophyllum commune Fr. and Molecular Phylogenegy within the Species, International Journal of Genetic Engineering, Vol. 13 No. 6, 2025, pp. 130-132. doi: 10.5923/j.ijge.20251306.09.
1. Introduction
- The diversity of living conditions of macromycetes and their trophic relationships give rise to different ecological groups of fungi. The inclusion of fungi in one or another ecological group does not depend on their systematic position. As a result, we can observe physiological and biochemical similarities during development among fungal groups that are phylogenetically distant from each other in terms of their way of life and nutrition [1].Biochemical adaptation, i.e., their adaptation to the nutrient environment and abiogenic factors, is of great importance in the formation of ecological groups of fungi. Since macromycetes are heterotrophic organisms, the nutrient environment is an important factor in providing them with the necessary nutrients. Fungi are divided into ecological groups: topical, saprotrophs, xylotrophs, coprotrophs, mycotrophs, corbotrophs, and mycorrhizal. In topical relationships, fungi use the trunk of a tree as a substrate. This, in turn, creates a favorable environment for the nutrition and reproduction of fungi [2].Schizophyllum commune is one of the fungi with high medicinal properties and widely used in pharmaceuticals and medicine. S.commune is a xylotrophic macrofungus belonging to the Basidiomycota division, Agaricomycetes class, Agaricales order, and Schizophyllaceae family. This species is one of the most widespread fungi in the world, occurring on all continents except Antarctica [3].Molecular identification of the medicinal fungus S.commune was carried out by a group of foreign scientists. The determination of the initial DNA sequence and molecular analysis of the fungus S.commune were carried out by foreign scientists, the determination of the rDNA isolates of the fungus and the analysis of their variability and the construction of the initial phylogenetic tree of the fungus were carried out by Japanese scientists [4]. The construction of more than 50 primer copies based on the unit consisting of 18S-5.8S-28S rRNA as a result of repeated transcription in the genome of S.commune was carried out by Indian scientists [5]. The search for similarities between the nucleotide and ITS sequences of the fungus 18S RNA was revealed in scientific research to search for small ribosomal RNA gene sequences in the Gen Bank [6]. Chinese scientists identified the 18S rDNA regions responsible for the schizophyllan biopolymer produced from the culture fluid of the medicinal S.commune using the ITS-1 and ITS-4 primers and mapped them to the NCBI sequences [7].As a result of our research, for the first time in Uzbekistan, the identification of the 1,5.8 S ribosomal RNA gene fragment from isolates of the medicinal fungus S. commune grown in liquid nutrient medium, the placement of the nucleotide sequence in the gene bank, and the molecular phylogeny within the species were carried out.
2. Materials and Methods
- The object of the study was the species S.commune, which is widespread in the Kashkadarya region. To carry out the scientific research, scientific trips were organized to different geographical regions of the Kashkadarya region, fungal samples were collected and brought to the laboratory. The fruiting bodies of the fungus S.commune were collected and isolated from the stems of poplar, walnut and local plum trees brought from the Shahrisabz, Guzar, Yakkabog and Kitab districts of the Kashkadarya region. Experimental mycology and microbiological methods were used to isolate a pure culture from the fruiting bodies of the fungus [8]. The samples collected from macromycetes were mycologically analyzed in the laboratory of the Department of Microbiology and Biotechnology of Karshi State University. The determination of the modern taxonomic position was carried out on the basis of electronic databases such as the international modern nomenclature of fungi Index Fungorum (ID:208403) and MycoBank (ID:208403).S.commune fungus was identified by molecular genetic methods in the Laboratory of Molecular Microbiology of the Institute of Microbiology of the UzRFA. The taxonomically significant DNA region of the culture was used for sequencing from ITS. For this, DNA was isolated from the culture and universal primers ITS-1 (5’-TCCGTAGGTGAACCTGCG-3’) and ITS-4 (5’-TCCTCCGCTTATTGATATGC-3’) were used [9].600 μl of the culture fluid of the fungus grown in liquid nutrient medium for 14 days was measured and placed in eppendorf tubes. It was centrifuged at 14,000 rpm and precipitated. Then the liquid was washed 3 times with distilled water, centrifuged and discarded. The cleaned and washed precipitate was dissolved with 600 μl of GTE (glucose-25 mM, tris-NSL- 20 mM and 10 mM EDTA). Lysozyme was added to the solution at a concentration of 2 μg/ml and kept in an ice container for 15 min. A final concentration of 2% SDS solution was added and incubated for 30 min at 37°C. The final concentration of the mixture was 0.3 M and kept on ice for 30 min. Then, they were centrifuged, and 600 μl of phenol (tris-saturated, pH-8.0) solution was added to them, mixed thoroughly for 2 minutes, and centrifuged at 13,200 rpm for 10 minutes. Then, the upper supernatant was measured, transferred to new test tubes, and once again, phenol solution was added to them, and centrifuged at 13,200 rpm for 10 minutes. In the next step, the upper supernatant was measured, transferred to new plastic test tubes, and a final concentration of 0.3M NaAc solution (pH-5.3) was added to it. Then, 2.5 volumes of cold 96% ethanol were added and placed in the freezer for 2 hours to precipitate DNA. Then, it was centrifuged at 13,200 rpm for 5 minutes and dried in an eppendorf thermoblock at 50°C for 20 minutes. In the final step, the DNA in the tube was dissolved in 60 μl of TE buffer [10]. The extracted DNA fragment was electrophoresed. The amplification and electrophoresis of the 1,5,8 gene region of the fungus were performed on a Biorad PCB, Main Board, CFX96/CFX96 Deep Well/CFX384 device [11]. Intraspecific molecular phylogeny of gene fragments of other state strains of S commune fungus was performed on the Mega X program [12].
3. Results and Discussion
- The strains isolated in the laboratory were initially registered in the collection "Macromycetes of the Southern Regions" at the Department of Microbiology and Biotechnology of Karshi State University. When naming strains, the first letter is the name of the collection, the second letter is the first letter of the authors' family, and the numbers indicate the registered position in the submitted collection (JE4, JE5, JE6).During the research, in order to fully analyze the S.commune fungal species, the nucleotide sequence of the 1,5.8 S ribosomal RNA gene fragment was determined. When compared using BLAST on the National Center for Biotechnology Information (nih.gov) online site, it was found to be 99% identical to the S.commune sample Sequense ID: MT647523.1 available in the gene database.According to the results obtained from the species sequence based on the ITS region of the fungal strains, no changes were noted in the species sequence, as the nucleotide sequences of JE5 and JE6 strains were almost identical. As a result, strain JE4 was registered in the NCBI (US National Center for Biotechnology Information) (https://www.ncbi.nlm.nih.gov/nuccore/PP837386, https://www.ncbi.nlm.nih.gov/nuccore/PP837387) EMBL-EBI European Nucleotide Archive (UK, Cambridge) and DDBJ Japanese DNA database under the ID numbers PP837386 ENA Browser (ebi.ac.uk) and PP837387 (ENA Browser (ebi.ac.uk).These analyses were performed to determine the identity of S.commune fungal strains and to construct an intraspecific molecular phylogenetic tree of strains occurring in different regions of the world based on the nucleotide sequences of these strains using the Mega X program (Figure 1).
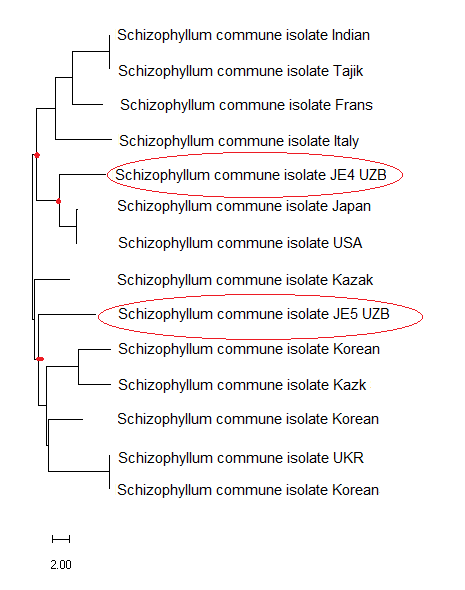 | Figure 1. Intraspecific molecular phylogeny of S.commune based on nucleotide sequences of strains JE4 and JE5 compared to strains isolated from different regions of the world |
4. Conclusions
- In conclusion, pure cultures of S.commune strains JE4, JE5 and JE6 were isolated for the first time, molecular identification of the strains was carried out with specific ITS-1 and ITS-4 primers, and based on the identification results, the isolates were registered in the National Center for Biotechnology Information (NCBI) database. During our studies, it was found that the JE4 strain of S.commune is close to the nucleotide sequences of the strains isolated in Japan, and the JE5 strain is close to the nucleotide sequences of the strains isolated in Korea. This, in turn, allows us to compare the nucleotide sequences of the strains with isolates isolated in different regions of the world and use these unique fragments in genetic engineering and biotechnology.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML