-
Paper Information
- Next Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Genetic Engineering
p-ISSN: 2167-7239 e-ISSN: 2167-7220
2024; 12(6): 120-122
doi:10.5923/j.ijge.20241206.10
Received: Nov. 20, 2024; Accepted: Dec. 12, 2024; Published: Dec. 16, 2024

Analysis of SSR Markers Associated with Yield Components of Soft Winter Wheat
Khamraev Nurbek1, Erjigitov Doston2, Yuldasheva Shakhnoza3, Qulmamatova Dilafruz4, Kushanov Fakhriddin5
1PhD in Biological Sciences, Khorezm Mamun Academy, Khiva, Uzbekistan
2Junior Researcher, Institute of Plant Genetics and Experimental Biology, Tashkent, Uzbekistan
3PhD Student, Khorezm Mamun academy, Khiva, Uzbekistan
4PhD in Biological Sciences, Institute of Plant Genetics and Experimental Biology, Tashkent, Uzbekistan
5DSc in Biological Sciences, Institute of Plant Genetics and Experimental Biology, Tashkent, Uzbekistan
Correspondence to: Khamraev Nurbek, PhD in Biological Sciences, Khorezm Mamun Academy, Khiva, Uzbekistan.
| Email: |  |
Copyright © 2024 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

In this article 18 local and foreign varieties of soft winter wheat were used in the study to analyze the SSR markers associated with yield components. It was observed that 5 out of 10 pairs of SSR microsatellite markers showed polymorphism, while the remaining 5 SSR markers were monomorphic. Winter soft wheat varieties showing polymorphism were involved as donors in selection work in order to create new varieties with high productivity and grain quality.
Keywords: Winter soft wheat, SSR markers, Yield components, DNA extraction, PCR and gel electrophoresis
Cite this paper: Khamraev Nurbek, Erjigitov Doston, Yuldasheva Shakhnoza, Qulmamatova Dilafruz, Kushanov Fakhriddin, Analysis of SSR Markers Associated with Yield Components of Soft Winter Wheat, International Journal of Genetic Engineering, Vol. 12 No. 6, 2024, pp. 120-122. doi: 10.5923/j.ijge.20241206.10.
1. Introduction
- Winter soft wheat (Triticum aestivum L.) is one of the food crops widely planted in the world and related to food security and social stability. However, its yield and quality are constantly challenged by various biotic and abiotic stresses [5]. Systematically evaluating the adaptability, disease resistance and stress tolerance of released wheat lines at molecular and genetic level is momentous to rationally distribute and apply them.Wheat production is strongly influenced by the size of cultivated land, climate change, agronomic methods, and different wheat varieties. The global cultivated land accounts for 14.2 million km2 or almost 10% of the Earth’s land area. However, factors such as aridity, soil erosion, and vegetation decline will degrade cultivated lands [7].The shrinking wheat cultivated area and the considerably negative influence of climate change may lead to decreased wheat production. However, global wheat production per annum increased by 1.0% [10]. The optimal crop management practices (including irrigation, fertilizing, pest/disease management, and weed control) [2] and the improvement of wheat varieties [1], have significant contributions in maintaining or enhancing wheat grain yield under adverse conditions.Yield stability under different environments relies on the compensatory of the main yield components, such as thousand-grain weight, kernel number per spike, and spike number per square meter. During the process of yield formation, the yield components perform differently under various environments. The adverse environments may hamper the specific yield components, and others may compensate for the loss of poor performance of specific components [12].A comprehensive understanding of the genetic gain in yield and yield-relevant traits is important for breeders and farmers to refine the directions of breeding programs and cropping cultivation. Three different approaches, the process-based models, statistical data, and field experiments, were applied to assess the effects of variety replacement on grain yield. All those approaches have specific advantages and limitations [6].The results from the field experiments are often more reliable and straightforward although more time and labor force are required [4]. In a previous study, the data from 2–3 year field experiments were used for analyzing the genetic gain in wheat yield using historical varieties in specific environmental conditions [13]. However, due to the limited number of experiments and varieties used, the results may not be robust [8].The use of yield as a selection criterion for the development of stress tolerant varieties is prohibitive in early generations. As well, drought stress does not happen predictably and evenly in the field. Therefore, plant breeders look for alternative methods such as marker assisted selection (MAS). Rapid advances in genome research and molecular technology have led to the use of DNA marker-assisted selection which holds promise in improving selection efficiency and expediting the development of new cultivars with higher yield potential [11].
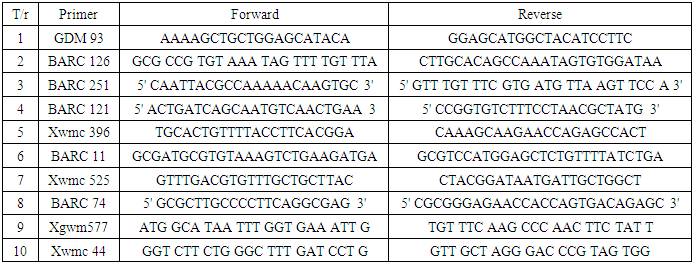
2. Materials and Methods
- Plant materials: 18 domestic and foreign varieties of winter soft wheat: 1-Chillaki, 2-Yaksart, 3-Bo‘zqal’a, 4-Yonbosh, 5-Do‘stlik, 6-Turkiston, 7-Polovchanka, 8-Vostorg, 9-Tobor, 10-Bezostaya-1, 11-Fortuna, 12-Kroshka, 13-Krasnodarskaya-99, 14-Pamyat, 15-Retezat, 16-MV-Nemere, 17-Kate-A1, 18-Konya 2002 varieties were used in the research.DNA Extraction and SSR marker assay: Varieties were grown under climate control at 24°C for 15 days to isolate DNA. The CTAB method was used to isolate genomic DNA from research samples. SSR markers specific for wheat were used to determine mutual genetic polymorphism between wheat varieties.PCR (hot-start wheat program) working mixture was prepared in a volume of 10 µl. The DNA concentrations of the samples were determined by gel electrophoresis by visual comparison with lambda (λ) phage DNA at a specific concentration (25 ng/µl) on a 0.9% agarose gel. Then, PCR amplification products were subjected to horizontal electrophoresis in 2.5% agarose gel at 100 V voltage for 90 minutes. Molecular weight marker is Invitrogen 50 bp DNA ladder (Cat. No. 10416014).
|
3. Results
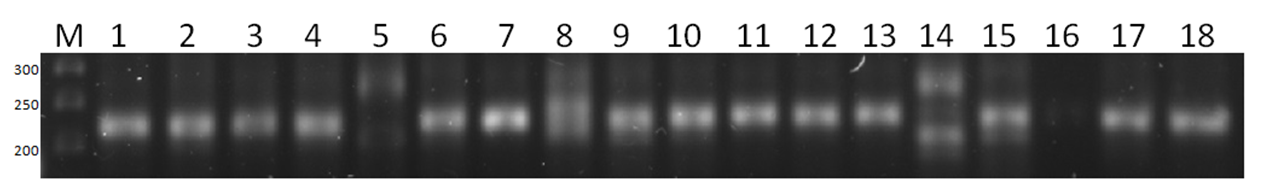
- It was observed that 5 out of 10 pairs of SSR microsatellite markers showed polymorphism, while the remaining 5 SSR markers were monomorphic. Winter soft wheat varieties showing polymorphism were involved as donors in selection work in order to create new varieties with high productivity and grain quality.Research samples were screened with microsatellite DNA markers using the polymerase chain reaction (PZP) method.In particular, the PCR analysis using Barc251 DNA marker, which is associated with productivity, allowed us to obtain the following results. According to this, the amplicons generated by this feature of the research samples were analyzed in the GelAnalyzer 19.1 bioinformatics program based on visual comparison with the weight marker with a molecular weight of 50 ng/µl. Based on the obtained data, 2, 4, 5, 6, 7, 9, 10, 2, 4, 5, 6, 7, 9, 10, 2, 3, 8, and 16 samples have 350, 240, and 270 nucleotide pairs, respectively, in samples 1, 3, 8, and 16. 11, 12, 13, 15 and 17 th samples are similar to each other it turned out to be (picture-1).
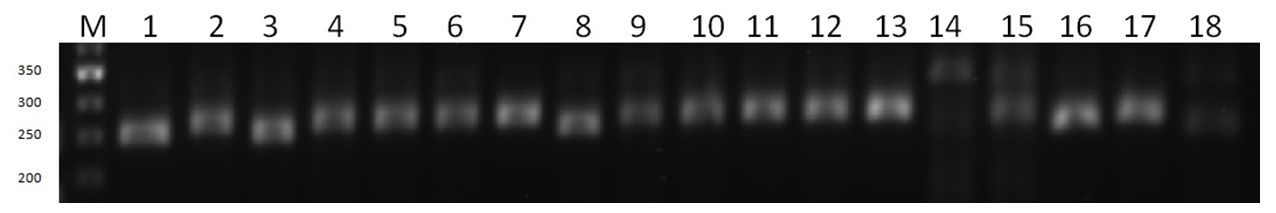 | Picture 1. Agarose gel electrophorogram of PCR analysis with BARC 251 microsatellite marker association used in in yield research |
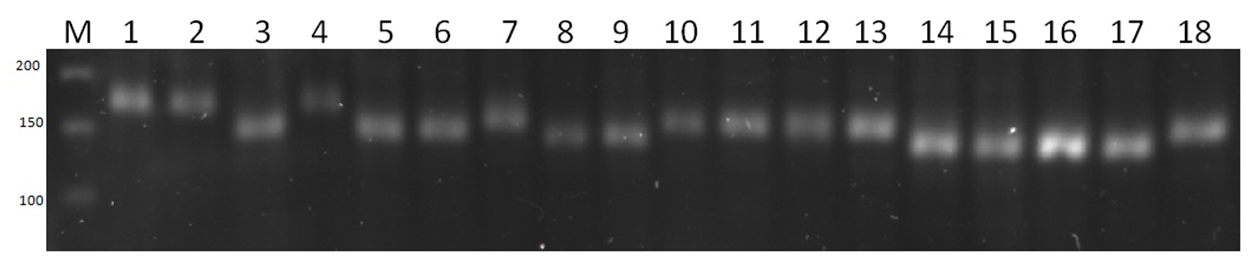 | Picture 2. Agarose gel electrophorogram of PCR analysis with the association Xwmc 396 microsatellite marker used in harvest index |
 | Picture 3. Agarose gel electrophorogram of PCR analysis with the association Xwmc 525 microsatellite marker used in 1000 grain weight |
4. Conclusions
- 18 local and foreign varieties of soft winter wheat were used in the study to analyze the SSR markers associated with yield components. It was observed that 5 out of 10 pairs of SSR microsatellite markers showed polymorphism, while the remaining 5 SSR markers were monomorphic. Winter soft wheat varieties showing polymorphism were involved as donors in selection work in order to create new varieties with high productivity and grain quality.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML