-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Genetic Engineering
p-ISSN: 2167-7239 e-ISSN: 2167-7220
2020; 8(1): 1-6
doi:10.5923/j.ijge.20200801.01

Computational Analysis of Single Nucleotide Polymorphism (SNPs) in Human MYOC Gene
Amged Mohammed Ibrahim, Afra M. Albakry, Nuha Widat Alla, Mona A. M. Khaeir, Hind. A. Elnasri
Department of Molecular Biology and Bioinformatics, College of Veterinary Medicine, University of Bahri, Khartoum, Sudan
Correspondence to: Mona A. M. Khaeir, Hind. A. Elnasri, Department of Molecular Biology and Bioinformatics, College of Veterinary Medicine, University of Bahri, Khartoum, Sudan.
| Email: |  |
Copyright © 2020 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Glaucoma is a disease that damages the eye’s optic nerve. It usually occurs when fluid builds up in the front part of the eye thus increasing the pressure within the eye and damaging the optic nerve. Among the causes of glaucoma is genetic polymorphisms of MYOC gene which can alter the myocilin protein and thus disrupting the regulation of the intraocular pressure which may lead to the disease. This study aimed to analyze nsSNPS in the Myocilin (MYOC) gene and the effect they may have on the protein function and structure. SNPs were obtained from the NCBI dbSNP database. The nsSNPs were further analyzed using 8 prediction tools namely GeneMANIA, SIFT, Polyphen-2, PROVEAN, SNPs & GO, PHD SNP, I-Mutant 3.0 and Project Hope. GeneMANIA results showed the association of MYOC gene with 20 other genes and mainly genes sharing the same protein domain. A total of 16 SNPs were predicted to be disease-associated using all software. Three SNPs were found to increase protein stability while 13 SNPs decreased the stability of the protein. In the current study, some SNPs that were previously reported to be associated with glaucoma were also found to be disease related using different software, while other new SNPs were predicted for the first time. In the future, these SNPs can clinically be tested to investigate their association with the disease.
Keywords: In silico analysis, MYOC gene, Glaucoma, Bioinformatics
Cite this paper: Amged Mohammed Ibrahim, Afra M. Albakry, Nuha Widat Alla, Mona A. M. Khaeir, Hind. A. Elnasri, Computational Analysis of Single Nucleotide Polymorphism (SNPs) in Human MYOC Gene, International Journal of Genetic Engineering, Vol. 8 No. 1, 2020, pp. 1-6. doi: 10.5923/j.ijge.20200801.01.
Article Outline
1. Introduction
- Glaucoma is a complex, heterogeneous ocular disorder with multi factorial etiology characterized by structural damage to the optic nerve, and commonly associated with relatively high intraocular pressure (IOP) [1-2]. It is a leading cause of irreversible blindness worldwide with ~20% of cases occurring secondary to other ocular or systemic diseases [2-4].Based on anatomical changes in the anterior chamber angle, primary glaucoma may be classified as primary angle closure glaucoma (PACG) or primary openangle glaucoma (POAG), which may be further subdivided into juvenile openangle glaucoma (JOAG) and adult onset POAG [1,5]. Glaucoma is a treatable disease if detected early; however, many patients are diagnosed during routine examinations or only following advanced field loss, as glaucoma is typically asymptomatic in the early stages. Therefore, the development of an accurate test for the detection of presymptomatic carriers at risk is important for the management of glaucoma.A family history of glaucoma is a well-known risk factor and hence genetic background is considered an important factor for the development of the disease [6-8].Several genes have been reported to be associated with primary glaucoma including myocilin (MYOC), WD repeat domain, neurotrophin 1, cytochrome P450 family 1 subtype [9-10]. To date, mutations in these genes account for only ~5% of patients with POAG, and the influence of mutations in these genes on patients with PACG remain controversial [11-12].The MYOC gene, is located on chromosome 1q24.3q25.2. Mutations in the gene are commonly found in juvenile or early adult patients with high IOP although mutation frequencies vary between ethnic groups [13].Bioinformatics is now playing a key role in different scientific areas. It involves computer sciences, mathematics, and statistics in order to analyze biological data that is being produced through the different sequencing techniques. Bio computing plays a key role in understanding the implication of genomic variations, especially single-nucleotide polymorphisms (SNPs), which represent the most frequent genetic variations in the human genome [14].SNPs are the single base change in coding or non-coding DNA sequence and are present in every 200-300 bp in human genome [15]. The nonsynonymous SNPs (nsSNPs) are the single nucleotide variations that affect the coding region of the protein and modify the mutated site-encoded amino acid, which may lead to a structural modification of the mutated protein, and may thus cause function alteration [15].The aim of the present study was to perform a computational analysis of the nsSNPs in the MYOC gene to identify the possible pathogenic SNPs and the effect they may impose on protein structure and function.
2. Materials and Methods
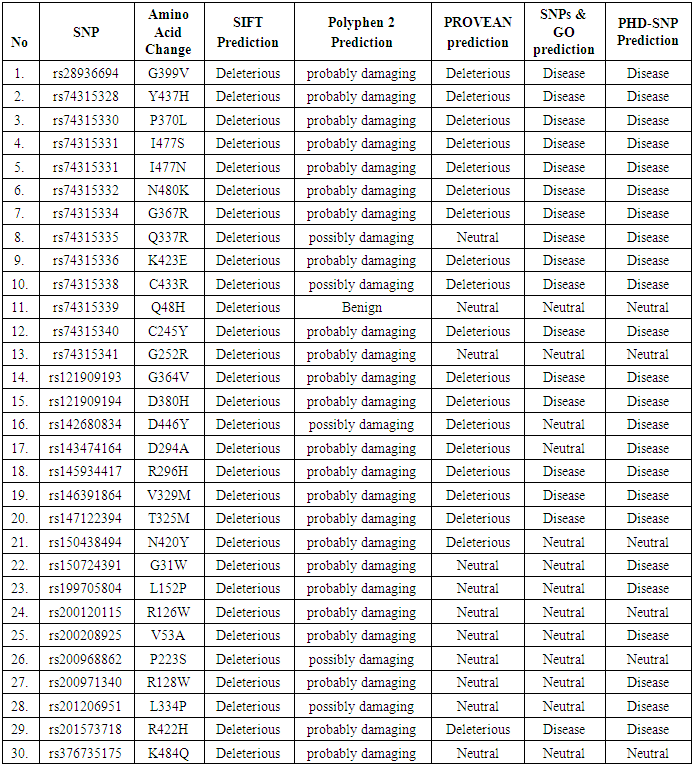
- SNPs in human MYOC gene data wereobtained from The National Center for Biotechnology Information (NCBI) dbSNP database during February 2020. The data obtained was further analyzed using various software.1- GeneMANIAGeneMANIA (http://www.genemania.org) is a web interface that helps predicting the function of genes and gene sets, can be used to find new gene members of a pathway or complex. MYOC gene name was entered as an input for GeneMANIA and the results were shown as a diagram showing the genetic interactions, pathways, co-expression, co-localization and protein domain similarity [16].2- Functional and structural analysis of SNPsSNPs retrieved from the dbSNP database were analyzed according to the scheme shown in Fig.1.
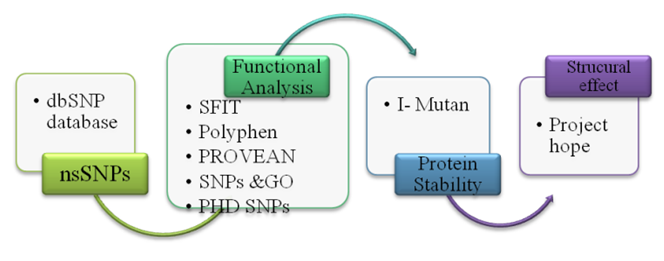 | Figure 1. Flow chart for SNP analysis |
3. Results
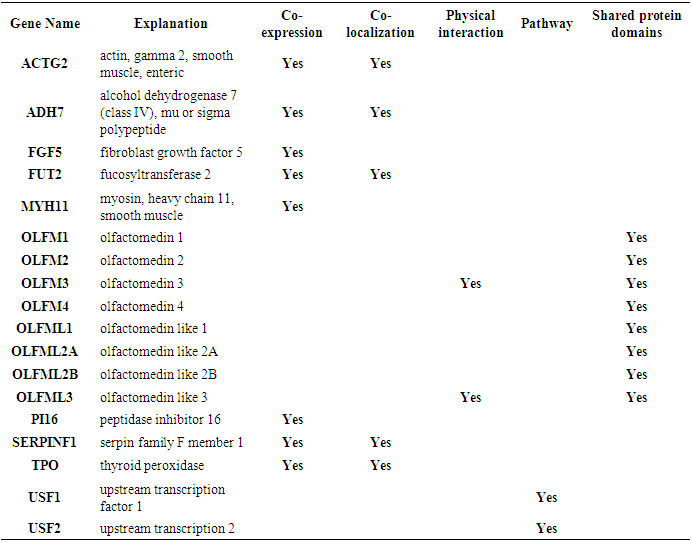
- Fig.2 shows the co-expression. physical interaction, shared protein domain between various gene and MYOC gene network. Eight genes OLFM1, OLFM2 OLFM3, OLFM4 OLFML1, OLFML2A OLFML2B, OLFML3 were having a shared protein domain. (Appendix 1). These genes are parlogs for MYOC and hence the shared domains.
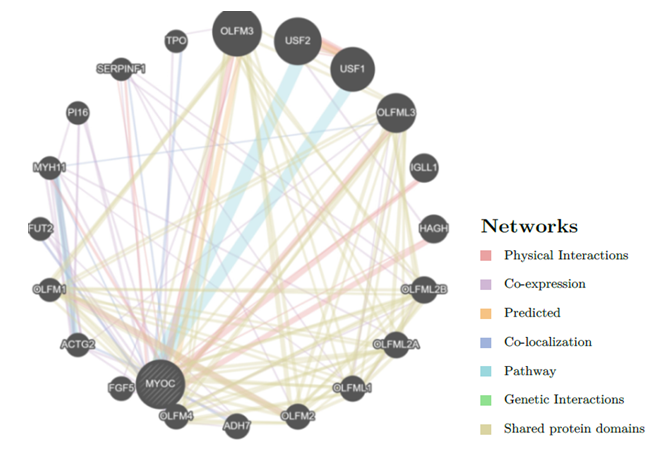 | Figure 2. GeneMANIA result for MYOC gene |
4. Discussion
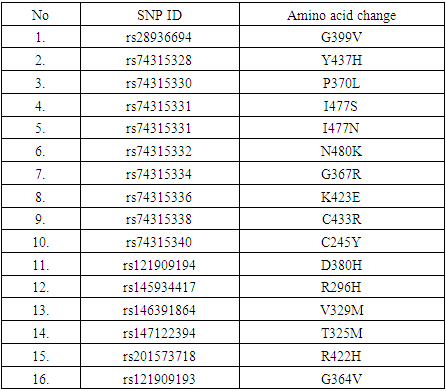
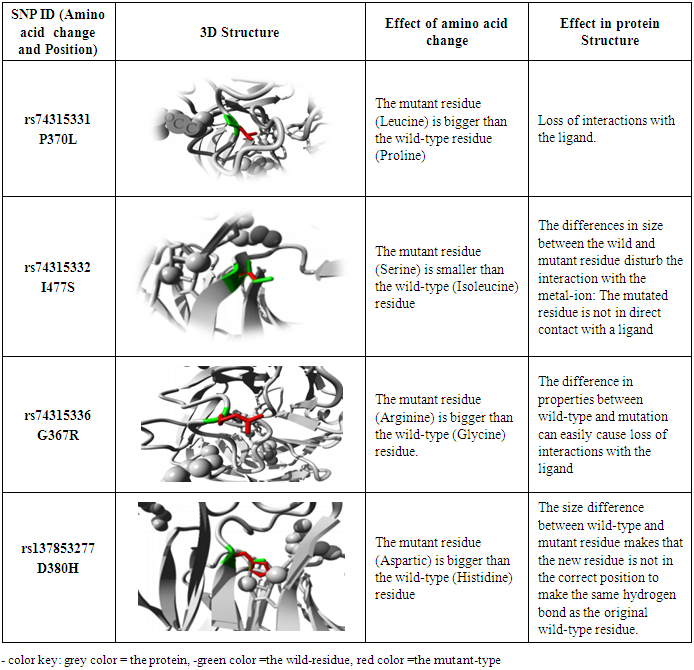
- In this study, investigation of nsSNPs in the MYOC gene was done using different computational software. A total of 16 SNPs were reported to be damaging using five different soft wares. A study carried among patients in Pakistan, showed the association of different SNPs in the MYOC gene and glaucoma, although with no statistical significance [23]. These SNPs (rs74315328, rs74315330, rs74315332, rs74315334, rs74315336, rs74315338 and rs121909193) were also confirmed in the current study to be disease related. Another SNP with rs74315341 has been reported among Caucasian and Brazilian population to be associated with glaucoma [24] but in this study this SNP was predicted to have a neutral effect using two software namely SNP and Go and PhD- SNP..Two SNPs were also reported to be disease related in this study and were also detected among Australian population (rs74315330, and rs74315334 [25]. Another SNP rs rs74315329 was reported as an important risk factor among Tasmanian population [26], but it has not been predicted in the present study. Another two SNPs (rs74315328 and rs74315331) were reported to be associated with hereditary glaucoma in the United states [27]. and were also confirmed in this study. Genetic defects can lead to an altered protein product which can be secreted into the extracellular matrix of the trabecular meshwork causing a severe form of autosomal dominant JOAG associated with very high IOP [9]. The effect of SNPs on protein structure can have different impacts such as increasing or decreasing its activity (as predicted by I Mutant) and hence affect the folding of the protein in the correct manner or affecting binding of the protein with specific types of ions or ligands as predicted by project hope- and can hence affect the function of the protein. Previous studies reported that mutated myocilin become tangled in the cell in its altered form [28].
5. Conclusions
- The current study showed that 16 nsSNPs are associated with glaucoma using various computational tools. These mutations can distort the protein stability or it’s binding with other ligands and thus affecting its function.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML