-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Genetic Engineering
p-ISSN: 2167-7239 e-ISSN: 2167-7220
2018; 6(1): 1-3
doi:10.5923/j.ijge.20180601.01

Analysis of Mutation Rate at 27 Y-STR Loci in "Grandfather/Father/Sons" Pairs from Uzbekistan Population
Kurganov Sardarkhodja, Axmedova Dilobar, Normatov Asilbek, Tosheva Dinara
Laboratory of Forensic Biological Examination of Human DNA, Republican Centre of Forensic Expertise, Republic of Uzbekistan
Correspondence to: Axmedova Dilobar, Laboratory of Forensic Biological Examination of Human DNA, Republican Centre of Forensic Expertise, Republic of Uzbekistan.
| Email: |  |
Copyright © 2018 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

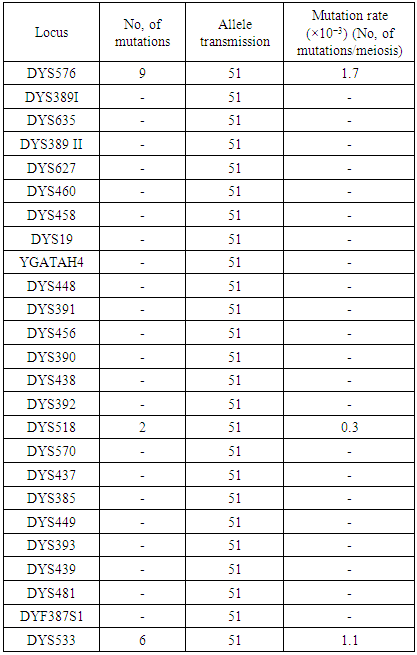
We analysed 68 samples from biological grandfathers, fathers and sons from Uzbek families were analysed in this study using Y-filer Plus assay, in order to estimate the mutation rate of 27 Y-STR loci in the population. Each families was previously confirmed by autosomal STRs testing (AmpFℓSTR® Identifiler Plus™ kit, Applied Biosystems) with paternity probability ≥99.99% and also confirmed mono-zygotic or dizygotic twins. Results showed that, In 17 grandfather-father-sons pairs, a total of 17 mutations were detected including 4 gains of single repeat mutations, 13 losses of single repeat mutations (note 1). An overall locus average of 1.2% rate of mutation was estimated for the 27 Y-STR loci. Our findings are encouraging and concur with previous studies showing that by 27-YSTR typing the discrimination power of male relatives could be considerably increased in comparison to every YSTR markers commonly used in forensic genetics.
Keywords: Y-chromosomal STRs, Y-STRs, Rapidly Mutating Y-STRs, Haplotypes
Cite this paper: Kurganov Sardarkhodja, Axmedova Dilobar, Normatov Asilbek, Tosheva Dinara, Analysis of Mutation Rate at 27 Y-STR Loci in "Grandfather/Father/Sons" Pairs from Uzbekistan Population, International Journal of Genetic Engineering, Vol. 6 No. 1, 2018, pp. 1-3. doi: 10.5923/j.ijge.20180601.01.
Article Outline
1. Introduction
- In the practice of forensic genetic examination, there are cases when the biological trace on physical evidence is left by an unknown male person. In such cases, along with the definition of the genotype of an unknown person, studies on the Y chromosome are performed on the STR loci of nuclear DNA. In most cases, these studies lead to a successful disclosure of the crime. But sometimes, despite the large number of samples examined, the suspect cannot be identified. In most such cases, DNA samples of individuals living in the same area are examined. To narrow the circle of suspects, calculate the lifetime of a common ancestor, through which it is possible to obtain information about related affinity and narrow the circle of suspects. Later, these results will be used to determine the relationship on the paternal line, where a reliable knowledge on mutation properties is necessary for correct data interpretation. When determining the degree of solution on the paternal line, if discrepancies between the child's father and other paternal relatives are not taken into account, population-specific mutation rates should be used to determine if this is a mutation or a true exception. There are at least 4500 STRs on the Y-chromosome, compared with many millions of potential SNPs. However, STRs mutate much faster, resulting in comparable mutation counts per individual: roughly one STR mutation and one SNP mutation every three to four generations [4]. Thus, STRs remain important markers for constructing fine-scale phylogenies.Y-STRs differ drastically in their mutation rates. Analyzed 186 STRs, and their mutation rates varied by two orders of magnitude, from 3.8 * 10−4 to 7.4*10−2 per locus per generation [1]. A similar range of variation was found among 67 STRs [3]. Most studies were based on family samples and capillary sequencing, but called STR-genotypes from full-genome datasets, using the same chromosomes as used for the SNP-based tree, and modeled STR mutations along the branches [4]. They demonstrated that motif size and the length of the longest uninterrupted tract (rather than total length of the locus) largely explain STR mutation rates. Tri- and tetranucleotide motifs underwent mainly single-step mutations, while dinucleotides frequently exhibited multistep signals. The authors estimated mutation rates for 700 STRs, including many slowly-mutating loci; thus rates varied by three orders of magnitude. Previously, we have already conducted work on the analysis of frequency of occurrence and mutation of alleles of 17 STR in the Y-filer (Applied Biosystems) in the Uzbek populations [2]. The aim of this study is to extend the existing data available regarding mutation rates of 27-YSTR(DYS576, DYS389I, DYS635, DYS389 II DYS627, DYS460, DYS458, DYS19, YGATAH4, DYS448, DYS391, DYS456, DYS390, DYS438, DYS392, DYS518, DYS570, DYS437, DYS385(a-b), DYS449, DYS393, DYS439, DYS481, DYF387S1(1-2) and DYS533) by analysing grandfather-father-son pairs samples from the Uzbek population.
2. Materials and Methods
2.1. Samples Collection
- Samples were collected from 17 (grandfathers, fathers and 2 sons) pairs unrelated males throughout Uzbekistan. During this study, samples were collected from paternity cases data tests involving males whose biological relationship were previously confirmed by autosomal STRs using AmpFlSTR Identifiler kit (Thermo Fisher Scientific), with a minimum paternity probability of 99.99%. All individuals signed the informed consent before participating in this study.
2.2. DNA Extraction
- Genomic DNA was extracted from peripheral blood and dried saliva samples using the phenol-chloroform-isoamyl alcohol method.
2.3. DNA Quantification
- After isolation, the quantity of genomic DNA of each sample was determined by quantitative real-time polymerase chain reaction (PCR) using the Quantifiler™ Human Male DNA Quantification kit (Thermo Fisher Scientific), which includes internal positive control to test for the presence of PCR inhibitors in the DNA extracts. Quantitative real-time PCR was performed on 7500 Real-Time PCR System (Applied Biosystems).
2.4. PCR Amplification and Detection
- To ensure successful amplification, using 0.75 ng to 1.5 ng of DNA was for each multiplex amplification. All thermal cycling was conducted on Applied Biosystems® GeneAmp® PCR System 9700 thermal cyclers. AmpFlSTRR Y-filer TM Plus PCR Amplification Kit (Thermo Fisher Scientific) PCR amplifications were performed as recommended by the manufacturer, although using half of the recommended reaction volume(12.5 μl).Separation and detection of the 17 Y-STR loci were performed using the 3130xl Genetic Analyser (Applied Biosystems) 16-capillary array system. Each samplewas prepared by adding 1 mL PCR product to 14 mL of Hi-DiTM formamide and 0.4 mL GeneScanTM-600 LIZTM internal size standard (Thermo Fisher Scientific).The sample run data were analyzed, together with an allelic ladder and positive and negative controls, using GeneMapper ID-X v1.4 (Life Technologies) software.
2.5. Statistical Analysis
- Comparison information of the sample data was generated using an in-house software program involving DNA-expert macros designed to check for allele sharing across all loci.
3. Results and Discussion
- A total of 68 samples from biological grandfathers, fathers and 2 sons from Uzbek families were analysed in this study using Y-filer Plus assay, in order to estimate the mutation rate of 27 Y-STR loci in the population (Table 1). In 17 grandfather-father-2 sons pairs, a total of 17 mutations were detected including 4 gains of single repeat mutations, 13 losses of single repeat mutations. An overall locus average of 1.2% rate of mutation was estimated for the 27 Y-STR loci.In our study, since no mutations were observed in DYS389I, DYS635, DYS389 II DYS627, DYS460, DYS458, DYS19, YGATAH4, DYS448, DYS391, DYS456, DYS390, DYS438, DYS392, DYS570, DYS437, DYS385(a-b), DYS449, DYS393, DYS439, and DYS481, DYF387S1(1-2) loci, the mutation rate of those loci was determined as zero (0).
|
4. Conclusions
- This study revealed important aspects of the mutational patterns of the 27 Y-STRs for the Uzbek population. There is a need to revise the criterion for exclusion in paternity testing taking into consideration the light of this and other studies. Larger number of samples should be further investigated in order to achieve more reliable conclusions especially regarding allele-specific mutations in the Uzbek population. Study of mutation rates of these markers is strongly encouraged for other populations in order to maximise the knowledge about such markers and also in order to define populations-specific mutation rates.Funding This work was supported by Ministry of Innovation Development of The Republic of Uzbekistan, Grant No. А-2-089+(А-2-056). Ethical approval This study was reviewed and approved by the ethics of the collective council of the Republican Center for Forensic Expertise (Republic of Uzbekistan).
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML