-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Genetic Engineering
p-ISSN: 2167-7239 e-ISSN: 2167-7220
2016; 4(1): 1-4
doi:10.5923/j.ijge.20160401.01

Validation and Enhance of a DNA Extraction Protocol in Palestinian Maize-Landraces
Yamen A. S. Hamdan
College of Agricultural Sciences and Technology, Palestine Technical University, Kadoorie (PTUK), Tulkarm, Palestine
Correspondence to: Yamen A. S. Hamdan, College of Agricultural Sciences and Technology, Palestine Technical University, Kadoorie (PTUK), Tulkarm, Palestine.
| Email: |  |
Copyright © 2016 Scientific & Academic Publishing. All Rights Reserved.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Assessment of genetic diversity is useful in plant breeding for different reason including the selection of parental combinations for developing progenies for specific traits. Development of modern plant breeding techniques has greatly facilitated wider use of a wealth of diversity from many sources including landraces. Molecular markers based on polymorphism of DNA sequence offer a powerful supplement to the morphological data currently used for variety protection, cultivar classification schemes, and estimation of the level of genetic diversity. However, extraction of enough DNA with satisfactory quality for molecular studies poses a challenge, most especially in developing nations. The objective of this study is to describe a DNA extraction protocol to extract high quality genomic DNA from leaf and seed of Palestinian maize landraces and to determines the suitability of extracted DNA for Polymerase Chain Reaction (PCR). In this study, we developed methods for isolating high-quality genomic DNA from leaves and seeds of maize crops with minor modifications. Extracted DNA will be done without the use of liquid nitrogen. DNA extracted from Palestinian maize landraces leaves as well as dry seeds samples yielded good quality DNA. The results revealed that the extracted DNA from the seed tissues is suitable for RAPD-PCR analysis.
Keywords: DNA extraction, Leaf, Seed, Zeamays, Landraces, PCR
Cite this paper: Yamen A. S. Hamdan, Validation and Enhance of a DNA Extraction Protocol in Palestinian Maize-Landraces, International Journal of Genetic Engineering, Vol. 4 No. 1, 2016, pp. 1-4. doi: 10.5923/j.ijge.20160401.01.
Article Outline
1. Introduction
- Maize (Zea mays L.) is the world's third most important in terms of area covered and is a multi-purpose crop for food, animal feed, biofuel, and raw material in the synthesis of a broad range of industrial products [1]. In Palestine, Maize is one of most important crops covering about 11,427.00 donums distributed all over the West Bank and Gaza strip. The annual production of maize is generally low and varies from year to year and from location to location, with a total production of only about 14874 tons [2].Assessment of genetic diversity is useful in plant breeding for different reasons including the selection of parental combinations for developing progenies for specific traits. Development of modern plant breeding techniques has greatly facilitated the detection of diversity from many sources including landraces.Molecular markers based on polymorphism of DNA sequence has been reported to provide information capable of determining the genetic constituents of plants and this is independent of environmental conditions [3]. Furthermore, molecular markers offer a powerful supplement to the morphological data currently used for variety protection, cultivar classification schemes, and estimation of the level of genetic diversity [4]. However, extraction of enough DNA with satisfactory quality for molecular studies poses a challenge, most especially in developing nations.Several studies have been carried out in recent years for molecular profiling of maize lines because morphological studies have been found to be influenced by environmental factors and not sufficiently stable as well as capable of producing misleading results [5]. Currently, the genetic diversity of plants has been assessed more efficiently after the introduction of methods that reveal polymorphism directly from the biochemical and DNA levels. Markers based on iso-enzymes and RFLP (Restriction Fragment Length Polymorphism) were the first molecular markers used in maize breeding programs. Then, markers based on polymerase chain reaction (PCR), such as random amplified polymorphic DNA or RAPD have been used in analysis of genetic distance in several plant species. Indeed, RAPD markers are commonly used because they are quick and simple to obtain, enabling genetic diversity analysis in several types of plant materials [6], such as natural populations, populations in breeding programs and germplasm collections. RAPD markers have been used in the analysis of genetic distance among segregant maize lines to predict the best crosses among lines for hybrid development, and to assess genetic diversity among collections of native maize.Generally, extraction of DNA from young leaf tissues is generally used for any genetic studies [7], [8]. Plant materials are among the most difficult for high quality DNA extractions because there are differences in structure and composition in different plants or different parts of the same plant [8]. For example, some plant parts contain high degree of tissue lignification and/or thick cell walls, while other cells contain more phenols; thus it is difficult to find unify and high quality DNA.This study describes the validation and enhance of a DNA extraction protocol to extract high quality genomic DNA from Palestinian maize leaf and seeds and to determines the suitability of both extracted DNA for polymerase chain reaction (PCR).
2. Material and Methods
2.1. Plant Materials
- Plant material consists of four Palestinian maize landraces including: PARC-1 obtained from Palestinian Agricultural Relief Committee (PARC); UAWC-10 from the Union of Agricultural Work Committees (UAWC); and TULK 1 as well as TULK 2 gotten from a specialized farmer in Tulkarm district.
2.2. Seed Germination for DNA Extraction
- Nine seeds of the four assumed cultivars were planted in plastic pots and placed in a growth chamber at a temperature of 24-30°C. Two weeks after germination, the second set of young leaves were collected from a three randomly selected seedlings of each assumed cultivars for DNA extraction. The collected leaf samples were put in well labeled paper bags and immediately upon collection, placed into a desiccator containing silica gel (Sigma-Aldrich). The ratio of silica gel to tissue should be not less than 10:1 by weight.
2.3. Reagents Employed
- 1. 1M Tris-HCL (pH 8.0, 9.5), 0.5 M EDTA (pH 8), 5.0 M NaCl; 3.0M sodium acetate (pH 5.2), CTAB (20%), chloroform and phenol:chloroform:isoamyl alcohol (24:24:1, v/v), β-mercaptoethanol.2. Modified CTAB extraction buffer: 0.1 M Tris-Cl (pH 9.5), 20 mM EDTA (pH 8), 1.4 M NaCl, CTAB (2%, w/v), β-mercaptoethanol (1%, v/v) (added to the buffer just before use).3. Pure cold (-20°C) isopropanol.4. 70% ethanol.5. Absolute ethanol.6. TE buffer: 10 mM Tris-Cl buffer (pH 8.0), 1 mM EDTA (pH 8.0).7. Enzyme: Taq DNA polymerase (Fermentas Inc.), Rnase a (Fermentas Inc.).8. Buffer: Taq DNA polymerase buffer.9. Nucleotides: dNTPs (G, A, T, C) and RAPD primer.10. TAE 5X: 24.2g Tris base, 5.71 ml Glacial acetic acid, 10 ml 0.5 M EDTA.11. Agarose gel; (12) 1X Gel red.
2.4. DNA Extraction from Maize Leaf Tissue
- 1. A dried leaf tissue (20 mg) were added to 2ml eppendorf tube containing three metal tungsten carbide beads and taped onto a vortexer and vortex on high setting for 30 seconds or until material is ground in a fine powder.2. A 500 μl extraction CTAB buffer were added to the ground tissue.3. The tubes were mixed by vortex for 30 s and incubated at 60°C in a water bath with shaking for 30 min, mix well by inversion each 5 minutes.4. Cool down tubes at room temperature for 5 min, then add 500 μl Chloroform to each tube, mix well by inversion, and centrifuge at 13000 rpm for 5 minutes.5. Aqueous layer (400 μl approximately) were transferred to a new set of tubes with 1 µl of 50 mg\ml RNase, mix well by inversion and incubate at 60°C for 30 minutes.6. Add 500 μl of absolute ice-cold isopropanol, mix well by inversion, and centrifuge at 13000 rpm for 10 minutes and discard the supernatant.7. Re-suspend DNA pellet in 500 μl high salt TE and incubate at 60°C for 30 minutes.8. Add 1 ml 100% ethanol to each tube, mix gently by inversion, and centrifuge at 13000 rpm for 5 minutes, then pour off the ethanol solution.9. Add 30 μl TE to each sample tube to re-suspend DNA, mix gently (to allow the bound DNA release into TE solution).
2.5. DNA Extraction from Maize Seeds
- 1. Seven seeds were grounded to a fine powder using pestle and mortar and (10 mg) transferred to a 2 mL eppendorf tube.2. A 800 μl modified CTAB extraction buffer were added to the ground tissue.3. The tubes were mixed by vortex for 30 second and incubated at 56°C in a water bath with shaking for 45 min.4. An equal volume (800 μl) of phenol, chloroform and isoamylalcohol (24:24:1, v/v) were added to each tube, mix well by inversion, and centrifuge at 13000 rpm for 15 minutes.5. Aqueous layer was transferred to a new set of labeled tube then add 800 μl of absolute ice-cold isopropanol, mix well by inversion, and centrifuge at 13000 rpm for 10 minutes and discard the supernatant.6. Pellets were washed with 800 μl l of 70% cold ethanol then tubes were centrifuged at 13000 rpm for 5min and left to dry completely at room temperature until no ethanol is remained.7. Re-suspend DNA pellet in 100 μl high salt TE with 2 µl of 5mg/ml RNase and incubate at 37°C for 60 minutes.8. For further purification an equal volume 100 μl of phenol, chloroform, and isoamylalcohol (24:24:1 V/V) were added to each tube, mix well by inversion, and centrifuge at 13000 rpm for 15 minutes.9. DNA were precipitated by adding 1/10 volume (20 μl) of 3M sodium acetate pH=5.2 and 2.5 volume (550 μl) of ice-cold ethanol the mixture were inverted gently and maintained for 30min at -20C and then centrifuged at 13000 rpm for 10min.10. The supernatant was discarded for each sample then the pellets were washed by adding 800 μl of 70% cold ethanol and the tubes were centrifuged at 13000 rpm for 5min and left to dry completely at room temperature until no ethanol is remained.11. Add 100 μl TE to each sample tube to re-suspend DNA, mix gently (to allow the bound DNA release into TE solution).
2.6. DNA Quality Evaluation by Agarose Gel Electrophoresis of PCR Products
- Quality of the DNA extracted from maize leaves as well as seeds was assessed by using electrophoresis of 2 μl on a 1.5% agarose gel. The genomic DNAs were used for amplification using a Random Amplified Polymorphic DNA primer (RAPD) OPX17: 5’- GACACGGACC -3’. Amplification of the DNA was performed in a thermocycler in the following manner: initial denaturation at 94°C for 1 min, followed by 47 cycles of denaturation at 94°C for 1min, primer annealing at 38°C for 45 sec. and extension at 72°C for 2 min, with a final extension at 72°C for 6 min.PCR-based amplification of the purified DNA was carried out in a (20 μl) -reaction mixture. The reaction mixture contained the following: 1x PCR buffer, 1.5 mM MgCl2, 0.2 mM dNTP, 0.6 μM of primer, 0.7 U of Taq DNA polymerase, and 50 ng of template DNA. The amplified products were separated on 1.5% agarose gel in 1×TAE buffer with 100 V in voltage for 30min and visualized on UV transparent.To prepare 1.5% agarose gel electrophoresis, 1.5g agarose powder were dissolved in 100 ml 1X TAE buffer and heated in microwave for 1:30 min, cool down at room temperature and add 3 μl of 1X Gel red, mix the gel by shacking and suspend in in a tray with 13 well comp and left to cool. DNA samples were loaded in wells by mixing 5 μl of extracted DNA with 1 μl of bromo blue dye.
3. Results and Discussion
- It is well documented that liquid nitrogen or freeze-drying is generally used for primary extraction [9], [10]. Here, my laboratory is the first to demonstrate that, DNA can be extracted from maize leaves and seeds without the use of liquid nitrogen which imply that considerable expense can be saved. Results revealed that, good quality of DNA was obtained via our proposed protocol mentioned above using fresh leaves and seeds. The quality of the DNA was tested by electrophoresis on a 1.5% agarose gel in which the genomic DNA fragment isolated had the same sizes and was over 20 kb (Figure 1). My result suggest that our improved DNA extraction method is successfully works since it produces high quality genomic DNA from Palestinian maize-landraces leaves as well as dry seeds. Adetumbi J.A. et al., [11] as will a Youssef. M. et al., [12] stated that good DNA quality is needed for molecular and genetic studies.
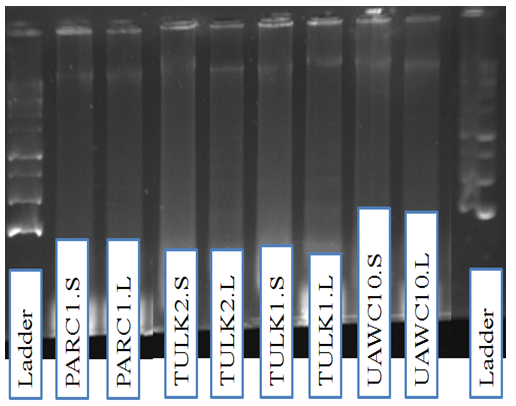 | Figure 1. Bands pattern of the DNA from leaves (L) and seeds (S) of Palestinian maize landraces |
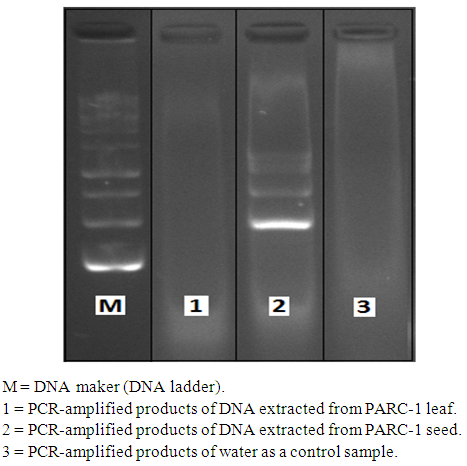 | Figure 2. RAPD-PCR amplification of Palestinian maize landrace (PARC-1) leaf and seed DNA using OPX17 primer |
ACKNOWLEDGEMENTS
- The author thanks the Palestinian Agricultural Relief Committee (PARC), the Union of Agricultural Work Committees (UAWC), for providing the seeds.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML