-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Genetic Engineering
p-ISSN: 2167-7239 e-ISSN: 2167-7220
2013; 3(2): 7-14
doi:10.5923/j.ijge.20130302.01
Transformation of Tropical Maize with the NPK1 Gene for Drought Tolerance
Rasha A. Omer1, 2, Jonathan M. Matheka1, Abdelbagi M. Ali2, Jesse Machuka1
1Department of Biochemistry and Biotechnology, Kenyatta University, P.O. Box 43844, Nairobi, Kenya
2Biosafety and Biotechnology Research Center, Agricultural Research Corporation, P.O. Box 126, Sudan
Correspondence to: Rasha A. Omer, Department of Biochemistry and Biotechnology, Kenyatta University, P.O. Box 43844, Nairobi, Kenya.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
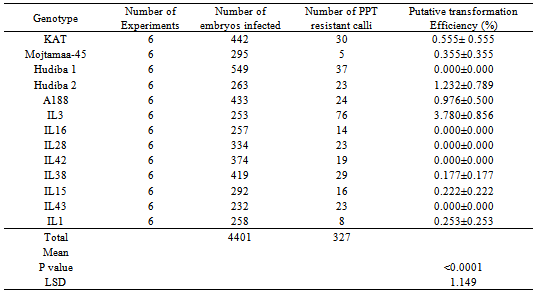
The objective of this study was to determine amenability of different tropical maize genotypes to Agrobacterium-mediated transformation with the NPK1 gene for conference of drought tolerance. To achieve this, immature embryos from thirteen maize genotypes (IL1, IL3, IL15, IL16, IL28, IL38, IL42, IL43, Hudiba-1, Hudiba-2, Mojtamma-45, A188 and KAT) were transformed by cocultivating with Agrobacterium tumefaciens strain EHA101 harboring the pSHX004 vector. Transgenic tussues were recovered on PPT (3 mg/l)-containing medium. Statistically significant differences (p<0.05) were observed between the genotypes with respect to transformation frequency (TF). Overall, IL3 was identified as the most amenable to transformation with a TF of 31.7% and proved to be superior to A188, which recorded a TF of 5.82%. Hudiba-2 was identified as the most transformable open pollinated variety (OPV) with a TF of 8.7% compared to that of 7.3% for KAT. IL1 and Mojtamma-45 proved to be poor responders to transformation with TFs of 2.5% and 1.7%, respectively. Putative transgenics were recovered from IL3, IL15, Hudiba-2, IL1, IL38, Hudiba-1, A188 and KAT. The frequency of regeneration of PPT resistant shoots varied from 6% for A188 to 100% for Hudiba-2. Stable integration of the transgene was confirmed by PCR. In conclusion we have demonstrated that tropical maize genotypes adapted to Sudan are transformable with the NPK1 gene.
Keywords: Tropical Maize, Agrobacterium-Mediated Transformation, NPK1, Drought Tolerance
Cite this paper: Rasha A. Omer, Jonathan M. Matheka, Abdelbagi M. Ali, Jesse Machuka, Transformation of Tropical Maize with the NPK1 Gene for Drought Tolerance, International Journal of Genetic Engineering, Vol. 3 No. 2, 2013, pp. 7-14. doi: 10.5923/j.ijge.20130302.01.
Article Outline
1. Introduction
- Transfer of desirable genes into agriculturally important plants has been achieved quickly and precisely using genetic engineering. When this has been attempted using particle bombardment, the transgenes integrate in a complex manner and in many copies. However, with Agrobacterium- mediated transformation, a few copies of the transgene are integrated in a simple pattern, resulting in stable transgene expression[1,2]. To date numerous reports have emerged indicating very low transformability of maize. The genotype seems to play a critical role in Agrobacterium-mediated maizetransformation with very few genotypes reported to be transformable. A188 is the most popular regenerable and transformable temperate genotypes[1,3]. Among the tropical maize genotypes, several genetic transformation experiments have employed CML126[4,5]. Apart from the genotype, the age and source of the explants is a critical success factor inAgrobacterium-mediated transformation. The only maize explants reported so far to be highly competent for Agrobacterium infection are immature embryos[6]. However these maize immature embryos show variations in their competence for Agrobacterium infection depending on their developmental stage[7]. Immature embryos of between 1.2 and 2.0mm have been the explants of choice in many Agrobacterium- mediated transformations experiments of maize[1,8–10].Agrobacterium-mediated transformation using the drought tolerance gene (NPK1) has been successfully achieved in temperate maize. Evaluation of the transgenic plants under drought conditions revealed a significantly higher photosynthesis rate than the non-transgenic controls [11]. The objective of this study was to evaluate the response of different maize genotype to Agrobacterium-mediated transformation. Maize immature embryo explants were transformed using Agrobacterium tumefaciens strain EHA101 was by the cocultivation method[8]. We report successful recovery of transgenic tissues using PPT as the selective agent. Further, we show successful regeneration of transgenic tropical maize plants that have stable transgene integration.
2. Materials and Methods
2.1. Plant Materials and Agrobacterium Strain
- Transformability was evaluated in eight Sudanese maize inbred lines (IL1, IL3, IL15, IL16, IL28, IL38, IL42 and IL43) and three commercial maize varieties (Hudiba-1, Hudiba-2 and Mojtamma-45) obtained from Sudanese Agriculture Research Corporation. Katumani (KAT) was used as a local open pollinated variety check, while A188 was used as the check inbred line. Seeds were planted in pots in the research field of the Plant Transformation Laboratory in Kenyatta University. A. tumefaciens strain EHA101 containing the standard binary vector pSHX004[12] was used in maize transformation. PSHX004 (Figure 1) contains the cauliflower mosaic virus (CaMV) 35S promoter (P35S) to drive both the bar selectable marker and the NPK1 gene. The vector system, pSHX004 in EHA101, was maintained on yeast extract peptone (YEP) medium containing 100mg/L spectinomycin (for pSHX004) and 50mg/L kanamycin (for EHA101). Bacteria cultures for weekly experiments were initiated from stock plates stored for up to two weeks at 4ºC
 | Figure 1. Map of the pSHX004 vector used in Agrobacterium-mediated transformation of maize |
2.2. Maize Transformation
- Agrobacterium cultures were grown for three days at 28°C on YEP solid medium amended with 100 mg/L spectinomycin, 50 mg/L kanamycin and 25mg/l Chloramphenicol. One loopfull (3 mm) of bacteria culture was scraped from the three-day old plate and was suspended in 5 ml of liquid infection medium (IM) supplemented with 100 µM acetosyringone (IM+AS) in a 50 ml centrifuge tube. The tube was fixed horizontally to a bench-top shaker and shaken on speed (~200rpm) for one hour at room temperature. This pre-induction step was carried out for all experiments. For infection, immature zygotic embryos from one ear were dissected to bacteria-free IM+ AS medium (2 ml) in 150x15mm sterile petri-plate (20 to 100 embryos). This was followed by the addition of 20ml of Agrobacterium suspension (OD550 = 0.3 to 0.4) to the embryos and left to stand in the dark for five minutes with occasional swirling. After infection, embryos were transferred to the surface of co-cultivation medium (CCM) and excess Agrobacterium suspension pipetted off the medium surface. Embryos were oriented with the embryo-axis side in contact with the medium (scutellum side up). Plates were then wrapped with aluminium foil and incubating in the dark at 20ºC for three days. After three days of co-cultivation, all embryos were transferred to resting medium (RM) and plates wrapped with parafilm before incubating at 28ºC in dark for 14 days. After 14 days on RM, embryos responding or not responding to callus induction were transferred to selection medium containing 1.5 mg /L PPT (SI) for 2 weeks. They were sub-cultured for two more 2-week passages on 3 mg /L PPT (SII) and wrapped with parafilm throughout selection. After the six weeks selection period, embryos surviving PPT selection were counted for calculation of Transformation frequency (TF). TF was computed according to Frame et al. (2006) as the percentage of the number of calli surviving PPT selection compared to the number of embryos infected.Acclimatization of regenerated plants was accomplished in soil in the glasshouse as reported for other tropical maize genotypes14. The number of regenerated plants was counted in order to compute the regeneration frequency (RF). RF was computed as the percentage of the number of regenerated shoots compared to the number of calli regenerating at least one shoot. Plantlets were maintained in the glass house till they matured and set fertile seeds.
2.3. Analysis of Transgenic Plants
- Leaf genomic DNA was prepared according the CTAB extraction method 15. The primers used to amplify the bar gene were forward primer5′-GTCTGCACCATCGTCAACC-3′ and reverse primer5′-GAAGTCCAGCTGCCAGAAAC-3. The PCRs were carried out in a 25μL solution comprising 10 ng of genomic DNA, 50 mM KCl, 10 mM Tris–HCl buffer (pH 8.8), 3 mM MgCl2, 0.1% (w/v) Triton X-100, 0.24 mM each dNTP, 1 U Taq DNA polymerase, and 0.16 μmol of each primer. Denaturation was performed at 94 °C for 1 min followed by 30 amplification cycles (94 °C for 30 s, 64°C for 2 min, and 72°C for 2 min) were separated in 0.8% (w/v) agarose gels. PCR product visualization and photography was achieved using the Genesnap image acquisition software from Syngene (Synoptics LTD). Transformation efficiency was computed on PCR positive events according to Frame et al. (2002) as the percentage of the number of PCR positive events per 100 Agobacterium-infected embryos.
2.4. Data Analysis
- Analysis of variance (ANOVA) was used to test the differences in responses to transformation frequency and efficiency using the Genstat for Windows (Discovery edition) statistical software. Means were compared using least significant difference (LSD) at the 95% confidence level. All percentage data was standardised using the square root transformation ((x+c)**0.5) before analysis.
3. Results
3.1. Selection of Stable Transformants
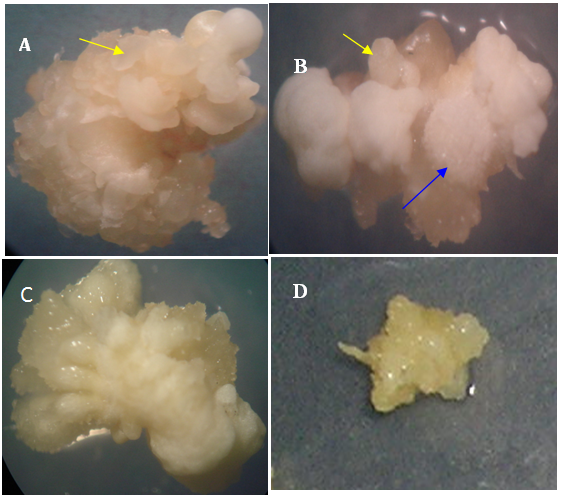
- After a period of co-culture and resting infected embryos responding or not responding to callus induction were transferred and maintained on selection media, containing PPT, for six weeks to select for stable transformation. All 13 genotypes used in this study were observed to initiate calli that survived and grew in PPT-containing media (Figure 2). However, the genotypes were different in the type of callus they initiated. Calli of IL3, Hudiba 2 and IL15 were observed to be white in colour, dry and compact. Moreover, friable embryos were clearly observable on the surface of these callus cultures (Figure 3). IL15 calli were observed to comprise of compact and friable sections and watery sections on their surface (Figure 3B). The inbred linesIL1, IL42, IL28 and Mojatamaa-45 were observed to induce a callus type that was watery and cream-colored in appearance (Figure 3C, D). These calli had a soft and spongy texture with no distinct embryo like structures observable on their surface. Rhizogenesis was also observed in some maize genotypes. Hudiba-1 and IL28 were particularly observed to germinate red, green and sometimes white pigmented roots during selection.
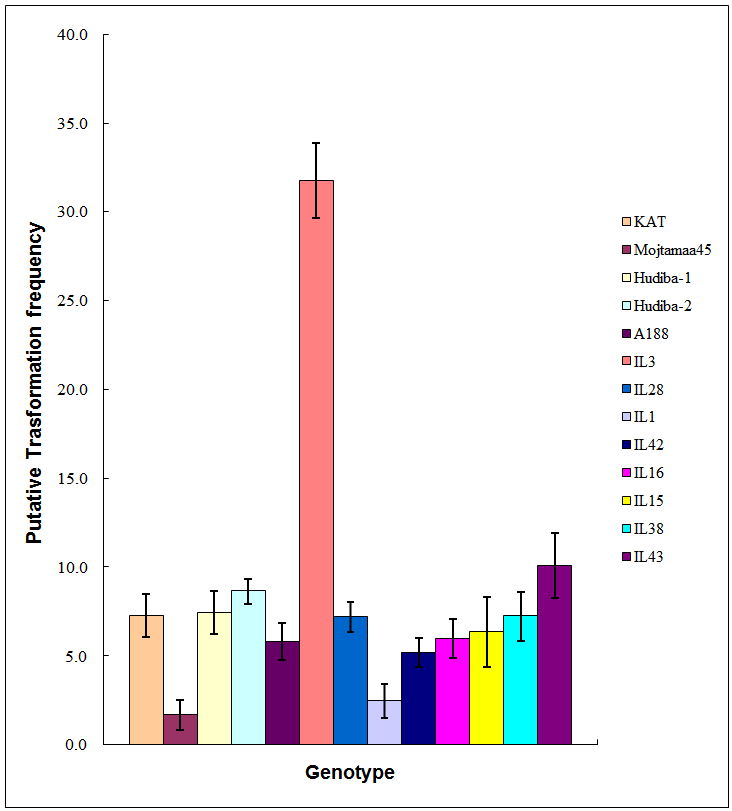 | Figure 2. Transformation frequency for various maize genotypes. Values are means of six replications and vertical bars are standard errors |
3.2. Transformation is Dependent on Genotype
- Transformation frequency (TF) was computed for all the 13 genotypes on which transformability was evaluated Figure 2. The average transformation frequency for all the 13 genotypes was 8.24%. The Open pollinated variety KAT and the inbred line A188 used as checks gave average transformation frequencies of 7.27% and 5.82% respectively. The highest average TF of 31.70% was observed in IL3 (Figure 2). The next highest TF was obtained in IL43, which ranged from 4.76 to 15.00% with an average of 10.09%. Hudiba-2 had a TF of 8.65% with a range of between 6.06 and 9.67%. The lowest TF ranged from 0.00 to 4.24%, with 1.69% as the average in Mojtamaa-45. The average TF obtained for IL1 was among the lowest obtained in this study (Figure 2). IL1 gave TF of between 0.00 and 6.06%, with an average of 2.47%. This was followed, in ascending order, by mean TFs of 5.19%, 5.99% and 6.34% produced by IL42, IL16 and IL15, respectively (Figure 2). The highest TF observed for IL42 was 5.56% while the lowest was 2.67%. IL16 had TF of 8.33% as the highest and 2.08% as the lowest, while the TF of IL15 ranged from 0.00% to 13.00%.Comparable average TFs were obtained for the genotypes IL28 IL38 and Hudiba-1. The Average TF recorded by IL28 was 7.13% with a range of 3.64% to 10.00%. IL38 had a TF of 7.24% with a range of between 2.70 and 12.12%. The TF of Hudiba-1 ranged from 3.43% to 11.54 % with 7.56 % as the average.Analysis of variance (ANOVA) showed that there were significant differences among the genotypes used in this study in transformation frequency (p<0.0001).Mojtamma-45 gave the lowest average transformation frequency, which was significantly lower (p< 0.005) than that of all the other genotypes. IL1 was inferior to other genotypes as its TF was observed to be the lowest (p<0.05). Differences were observed in TF between IL3 and the other genotypes, in that the TF produced by IL3 (31.70%) was found to be significantly higher (p<0.0001). Significant differences were observed among the OPVs used in this study. The local check OPV (KAT) was observed to produce a significantly higher TF (p<0.005) than that of Mojtamaa-45, a Sudanese OPV. KAT, Hudiba-1 and Hudiba-2 produced almost comparable TFs. No significant differences (p>0.05) in TF were found among them. Variations were found among Sudanese OPVs with respect to transformation frequency. Mojtamma-45 gave a significantly lower TF (p<0.005) compared to other Sudanese OPVs. ANOVA also showed significant differences between Sudanese inbred lines and the A188 genotype used as the check inbred line. The Putative TF of 31.70% produced by IL3 was found to be significantly higher (p <0.0001) than that of A188 (5.82%) as well as the other Sudanese inbred lines (p<0.0001). However, no significant differences (p>0.05) were found between A188 and the rest of the Sudanese inbred lines. IL1 gave the lowest average of TF. A significant difference (p <0.0149) was observed between IL1 and A188 with respect to TF. Moreover, significant differences in TF (p< 0.005) were found between IL1 and all other Sudanese inbred lines.
3.3. Transgenic Plant Regeneration is Influenced by Genotype
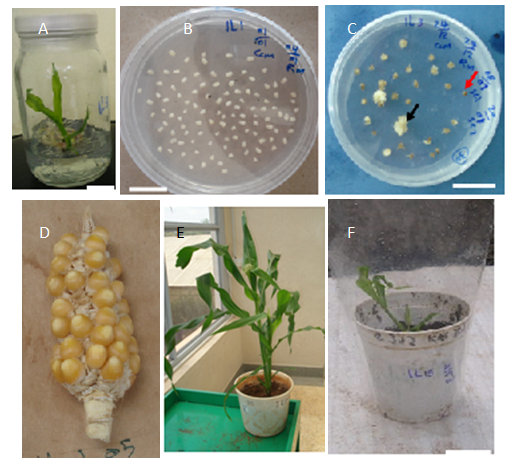
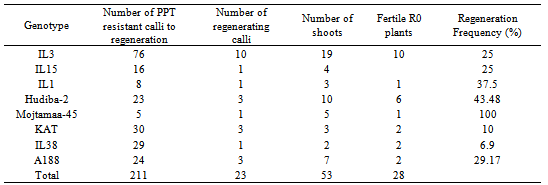
- Putative transformation efficiency (TE) was calculated for all the maize genotypes on which transformation was attempted. Results indicate that most of the Sudanese varieties were particularly incompetent in establishment of plants. This is reflected by the low putative TE values they recorded, that are as low as 0.00% (Table 1). Only eight genotypes produced plantlets they grew to maturity. The standard control in bred line (A188) and the local check OPV (KAT) recorded TE of 0.98 and 0.56% respectively. The highest TEs were observed in IL3 and Hudiba-2. IL3 averaged 3.78% in TE, with a range of 2.70% - 5.46%. Hudiba-2 was observed to yield an average TE of 1.23% with a range of between 0.00% and 4.17%. IL15 and IL1 recorded TEs that were lower than those of the control genotypes (Table 1).ANOVA shows that there was an influence by the genotype on the observed T.E. response (Table 1). The T.E of IL3 was significantly higher (p<0.05) than those of all the other genotypes, including A188. The best performing OPV with regard to T.E. was Hudeiba-2, whose efficiency of transformation was significantly higher (p<0.005) than that of other OPVs.PPT surviving calli from all the genotypes were placed on maturation (Regeneration 1) media and cultured for 14 days in the dark. During this period, somatic embryos of A188, IL3, IL15, IL1, IL38, Mojtamma-45, Hudiba-2 and KAT calli matured, that is they formed a scutellum (that later turned white), a coleoptile-like structure (that turned green) and a rootlet. Calli of the other genotypes had no observable response to maturation.Mature calli transferred to shoot induction media (Regeneration I) started greening 2-5 days after transfer to light. From majority of the calli, shoots and roots regenerated on R1 (Figure 4C) and transfer to RII was therefore unnecessary. The calli of Hudiba-1 and IL28 cultured on R1 responded to maturation by forming roots, which were mostly red pigmented. Calli of Mojtamaa-45, IL1, IL43 and IL28 had no other observable response to regeneration except slight greening.Since all the genotypes produced PPT resistant calli, regeneration was attempted for all the genotypes. However, not all genotypes could regenerate plants. Regeneration was achieved only in IL3, IL15, and IL1, IL38, Hudiba-2, Mojtamma-45, A188 and KAT. These genotypes regenerated 19, 4, 3, 2, 10, 5, 7 and 3 plants, respectively (Table 2). Mojtamma-45 had the highest regeneration frequency (Table 2). However regenerated shoots were weak and delicate in growth resulting in the death of some of them. Moderately high regeneration frequency of 43%, 37% and 29% were observed for Hudiba-1, IL1 and A188, respectively. The lowest regeneration frequency was recorded by IL38 (Table 2).
|
|
3.4. Bar and NPK1 Transgene was Stably Integrated into the Maize Genome
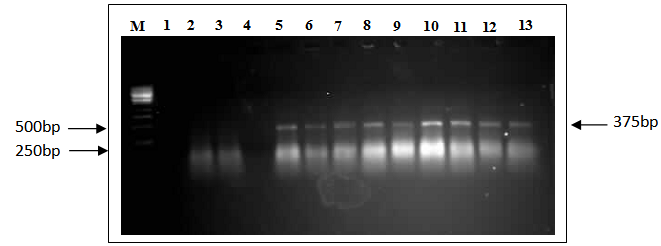
- Seedlings of putative transformants were assessed for the presence of the bar gene. Gel electrophoresis of genomic DNA extract from the leaves of the seedlings revealed minimal degradation during extraction. The presence of bar gene in the putative transformed R0 and R1 plants was detected by PCR amplification of the 375 bp fragment in the DNA extracts from transgenic plants. This fragment represents the bar gene element on the pSHX004 plant expressible cassette. DNA from a non-transformed Sudanese maize genotypes was used as the negative control whereas pSHX004 plasmid DNA was used as the positive control. There was amplification in the sample containing the plasmid DNA and no amplification occurred in the negative controls (Figure 5).
4. Discussion
- Nine inbred lines and four OPVs were studied in relation to their response to Agrobacterium-mediated transformation. Necrosis and cell death was observed in some embryos at the co- cultivation stage. IL1, IL16 and Mojtamma-45 embryos were especially observed to respond to Agrobacterium co-cultivation with necrosis and arrested growth. Agrobacterium induced necrosis has been reported in maize under co-cultivation conditions[16]. Factors that influence the degree of necrosis include explant age, preculture period, bacterial density and infection duration[17,18].It was observed that all the inbred lines and the OPVs are transformable since some callus events survived PPT selection. Survival of the callus was because they harbored the bar gene which codes for the phosphinothricin acetyltransferase (PAT) enzyme that acetylates PPT. Acetylated PPT is no longer inhibitory to glutamine synthase[19]. Untransformed cells exposed to PPT die as a result interruption of protein synthesis by phosphinothricin (the active component of PPT). Phosphinothricin (PPT) is a potent inhibitor of glutamine synthase. Inhibition of glutamine synthase leads to a rapid accumulation of ammonia in the cells leading to cell death[19].Results indicate that the TF response varied with the genotype used. Similar findings were reported by Ombori and coworkers[5] who observed genotype specific responses in TF when Agrobacterium was used to transform tropical maize genotypes with the bar gene. A188 a temperate inbred line, has been proven to be an effective Agrobacterium host and performs well in culture. Therefore it is used widely to extend the range of maize genotypes susceptible to Agrobacterium[20,21]. Results indicate that A188, used a standard check inbred line, produced a putative TF of 5.8%. A TF of 5.5% was obtained for A188 by Frame and coworkers[8] utilising the Agrobacterium strain EHA101 harbouring pTF102 binary vector construct. However, TFs of between 5% and 30% have been realised in A188 using Agrobacterium harbouring a superbinary vector[22]. The average TF obtained for the Sudanese Inbred lines was 8% and ranged from 2.5% for the IL1 to 31.7% for IL3. A high TF was obtained for IL3 despite the recalcitrance of tropical maize, especially in bred lines, to Agrobacterium-mediated transformation. In fact, the putative TF of IL3 was observed to be higher than that of the standard check, A188. IL1 on the other hand is inferior in TF to A188 and indeed all other genotypes evaluated.Transformed calli were observed to regenerate between 1-3 shoots per callus, indicating a relatively high capacity of PPT resistant calli to regenerate shoots. Regeneration frequencies of PPT resistant shoots varied from 100% for Hudeiba-2 to 6% for A188. Tissue culture response and plant regenerability has been shown to be genotype specific[23,24] and therefore under complex genetic control. Separate genes have been shown to govern these processes[20,25]. However, not all PPT resistant events were regenerable. For example, IL16, IL42, IL28, IL43 and Hudeiba-1 had some callus surviving PPT selection but no shoots could regenerate from them. These genotypes recorded TE of zero. Regenerability has been found to be genotype-dependent[26]. In conclusion, this study has demonstrated that transformation using Agrobacterium tumefaciens is possible for a number of tropical maize genotypes. Successful stable transformation of Sudanese in bred lines (IL3, IL1, IL15 and IL38) and OPVs (Hudiba-2 and Mojtamaa-45) using the NPK1 gene was achieved. A total of 28 transgenic plants were obtained from these genotypes. The inbred line IL3 identified in this study as highly transformable extends the pool of tropical maize genotypes that can be utilized in quick insertion of genes of agronomic importance through genetic engineering.
ACKNOWLEDGMENTS
- The work done at the Plant transformation Laboratory at Kenyatta University under ASARECA funding. We would like to thank Professor Kang Wang from IOWA State University for providing the gene construct. We acknowledge the Agricultural research corporation of Sudan for providing the maize germplasm used in this study.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML