-
Paper Information
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Genetic Engineering
p-ISSN: 2167-7239 e-ISSN: 2167-7220
2011; 1(1): 6-13
doi: 10.5923/j.ijge.20110101.02
Genetic Modification of Pichia Pastoris for Production of Propylene Glycol from Glycerol
Guillaume G. Barbier , Jacob L. Ladd, Ellen R. Campbell
The Nitrate Elimination Company, Inc. (NECi), 334 Helca St., Lake Linden, MI, 49945, USA
Correspondence to: Jacob L. Ladd, Ellen R. Campbell , The Nitrate Elimination Company, Inc. (NECi), 334 Helca St., Lake Linden, MI, 49945, USA.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
Biodiesel is emerging as a major renewable energy resource. Glycerol is byproduct of biodiesel production, which cannot be directly used as fuel. Using Pichia pastoris as aerobic yeast cell factory, we engineered pathway to convert glycerol to propylene glycol (PG; 1,2-propanediol, 1,2-dihydroxypropane), a fuel additive and commodity compound. PG biosynthetic pathway was designed with unique combination of 3 enzymes: Escherichia coli methylglyoxal synthase and alcohol dehydrogenase; and Pichia ofunaensis glycerol dehydrogenase. Engineered genes were successfully integrated in yeast genome and actively expressed. Analysis, by HPLC of shake flask and fermenter broths of glycerol-grown cells, demonstrated engineered yeast produced PG. In fermenter, highest PG concentration was 0.11 g/L. When engineered P. pastoris was grown on biodiesel crude glycerol as sole carbon source, PG was detected. Basically, this is a proof of concept to demonstrate that Pichia was engineered to produce PG from glycerol.
Keywords: Genetic Engineering, Glycerol, Biodiesel, 1,2-Propanediol, Propylene Glycol, Fuel Additive, Pichia Pastoris
Cite this paper: Guillaume G. Barbier , Jacob L. Ladd, Ellen R. Campbell , "Genetic Modification of Pichia Pastoris for Production of Propylene Glycol from Glycerol", International Journal of Genetic Engineering, Vol. 1 No. 1, 2011, pp. 6-13. doi: 10.5923/j.ijge.20110101.02.
Article Outline
1. Introduction
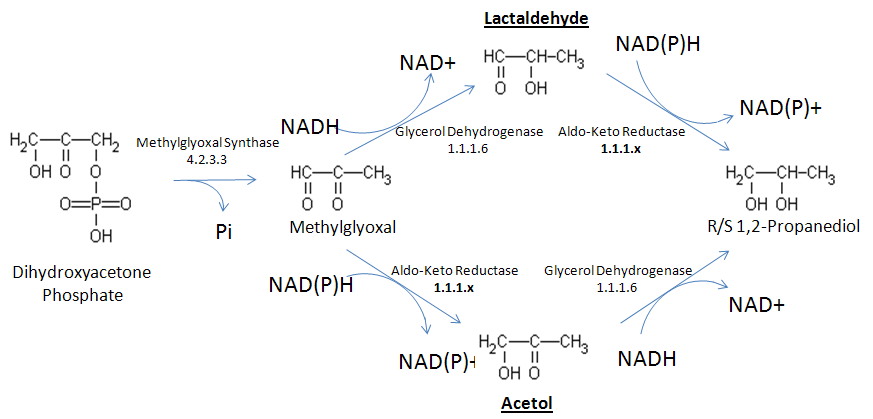
- With decline of natural fossil fuel reserves of the world, development and industrial production of renewable energy has expanded to supply increasing world energy demand. Perhaps the most successful efforts in this direction so far have been in production of biodiesel from crop plant resources. In biodiesel technology, plant lipids are transesterified with methanol, leading to formation of three long chain hydrocarbon fatty acid methyl-esters, and a molecule of glycerol. The fatty acid esters are the fuel in biodiesel, and glycerol is a byproduct that cannot be used as fuel. It is estimated that for every 9 kg of biodiesel produced, 1 kg of crude biodiesel glycerol (crude glycerol) is generated. The amount of byproduct crude glycerol from biodiesel production was about 200 million pounds for 2006, and should be reaching 1.4 billion pounds by 2011 in the US alone[1]. The annual glycerol consumption in the US is about 400 million pounds per year, so production is now well exceeding demand.Two billion pounds of Propylene Glycol (PG) are produced yearly in the US by petroleum-based processes, and the annual market growth is about 4%. The petroleum-based PG production involves chemical processes that are highly energy consuming and relies on a supply of fossil fuel-based propylene. Genetic engineering of Escherichia coli and the yeast Saccharomyces cerevisiae for aerobic production of 1,2-propanediol from glucose have been described[2-4], and a patent application filed[5].Glucose is assimilated and undergoes glycolysis. The entry point of the PG pathway is dihydroxyacetone phosphate which is metabolized to methylglyoxal (MG). From there, two pathways are found to produce PG, depending on the sequence of reduction of oxo groups of MG (Fig. 1). In one route, also called the lactaldehyde-forming pathway, the 2-oxo group is first reduced to form lactaldehyde, and then the 1-oxo group of lactaldehyde is reduced to form PG. The alternative route, known as the acetol-forming pathway, is to reduce the 1-oxo group first, which leads to acetol, and then reduce the 2oxo of acetol to yield PG.
2. Methods
2.1. Gene Synthesis
- The synthetic PG pathway cassette containing the mgsA, yqhD and glydh gene cassettes was contiguously synthesized by Genscript (Piscataway, NJ) according to our precise specifications and was delivered in a pGAPZ plasmid (Invitrogen, Carlsbad, CA) from which the “endogenous” Pgap was previously removed by restriction digestion (pPG1, see Fig. 3). Each gene cassette was designed with the following parts: strong constitutive glycerol aldehyde phosphate dehydrogenase Pichia promoter sequence (Pgap), a consensus ribosome binding site (RBS), the gene of interest-codon-optimized for expression in Pichia, transcription termination from Pichia alcohol oxidase 1 gene (AOX1 TT) and restriction sites (see Fig. 3A).
2.2. Plasmid Transformation and Selection
- The pPG1 plasmid (Fig. 3B) was transformed into E. coli Novablue electrocompetent cells. Clones were selected on LB-LS plates containing 50 µg/L Zeocin®, which is the antibiotic corresponding to the resistance gene in the pGAPZ plasmid. About 20 Zeocin® resistant E. coli clones were randomly picked from the plates and screened by colony PCR for the presence of the genes using gene specific primers (35 cycles, denaturation: 94℃, annealing: 57℃ extension: 72℃). The gene specific forward and reverse primers for mgsA were:5’-CGATCACTGCAAACAAATGC-3’and 5’-CGCGAGATAACGCTGATAATC-3’.The gene forward and reverse specific primers for yqhD were: 5’-CCGCATTCTGTTTGGTAAAG-3’and 5’-GGTGTAGGTATAAACCGGATC-3’.The gene forward and reverse specific primers for glydh were:5’-CAATCAGGTAGCCATCTCTG-3’and 5’-ATCTTGACGTCGTGACTACTC-3’.PCR products for each of the 3 genes were analyzed by agarose gel electrophoresis. One PCR positive E. coli cell line (E. coli pPG1), thus containing the pPG1 plasmid, was grown in 100 mL LB-LS medium with 50 µg/mL Zeocin® (37℃, 16 h, 220 rpm). From the harvested cells, the pPG1 plasmid was extracted and purified using Promega Wizard DNA prep (Madison, WI).
2.3. HPLC Analysis of Pichia Culture Spent Broth
- Spent cell broths were concentrated 5 fold using a freeze dryer, filtered, centrifuged (3 min, 10,000 rpm) and then analyzed with a Waters Corporation HPLC system equip- ped with a Waters 410 differential refractometer (Milford, Ma). The column was a Bio-Rad Aminex HP-87X (Bio-Rad, Hercules, Ca), and the mobile phase, 0.025 M sulfuric acid, was pumped at 0.3 mL/min flow rate. The column temperature was kept at 40℃ and we used a 50 µL injection loop. Results were matched against wild type Pichia extracts and spent broths, and positive samples were re-run and spiked with a known amount of PG standard. Under these conditions, PG retention time was 32.75 min. A standard curve was made using the PG standard to quantify and determine the detection limit of the product.
2.4. Pichia Extraction and Enzyme Activity Assays
- For each cell lines, cells were centrifuged (10 min, 20,000 g, 4℃) and resuspended in breaking buffer (50 mM phosphate, pH 7.0) to a concentration of 50-100 OD600 nm. Cells were disrupted using a Mini-Bead-Beater (Biospec Products, Bartlesville, OK): 1 mL of cell suspension was mixed in a micro-centrifuge tube with 0.5 mL of 0.5 mm zirconium-coated glass beads. Samples were then disrupted by processing them for 1 min with the Mini-Bead-Beater, and placed on ice for 2 min. The cycle was repeated 3 times, and tubes were centrifuged 10 min, 20,000 g, 4℃. Supernatant was collected and designated the Centrifuged Crude Extract (CCE).CCE (10-100 µL) of Pichia-wt and Pichia-PG1 were used to assay enzyme activities at room temperature (~22℃). MSG activity was monitored by following appearance of MG by the DNPH method as previously described (Hopper and Cooper, 1972). One unit of MGS activity was defined as the amount required for the formation of 1 nmol of MG per minute under assay conditions. YqhD methylglyoxal reductase activity was assayed by following NADPH disappearance at 340 nm. Crude extract (100 µL) was incubated in assay buffer, which contained 50 mM Tris-Cl (pH 7.7), 10 mM MG, 1 mM EDTA and 1 mM 2-mercaptoethanol. After incubation at 37℃ for 2 min, 100 µL of NADPH at 5 mM was added, and the reaction was followed on a spectrophotometer at 340 nm. One unit of YqhD MG reductase was defined as the amount required for the disappearance of 1 nmol of NADH per minute under assay conditions. GlyDH acetol reductase activity was determined by observing the disappearance of NADH at 340 nm as previously described[16]. One unit of GlyDH acetol reductase was defined as the amount required for the disappearance of 1 µmol of NADH per minute under assay conditions.
2.5. Fed-Batch Fermentation
- Pichia-PG1-1 cell line was grown in a one-liter fermenter run for 50 h, using a standard in-house protocol developed for Pichia[8]. The carbon source was ACS grade glycerol. The fermenter was a BioFlo3000 (New Brunswick Scientific, Edison, NJ) with a vessel of 1.25 L. The fermenter was interfaced to a computer using the NBS-BioCommand software. The fermenter was operated at 30℃, the pH was maintained at 4.9 by addition of 28 % NH4OH, and the dissolved oxygen level was kept above 35 % by using the DO cascade loop of the fermenter. The glycerol introduced was monitored using a balance (APX 6001, Denver Instruments) interfaced to a computer via Winwedge Pro software (Tal Technologies, Philadelphia, PA).First, cells were grown in batch mode for about 24 hours, and when all the glycerol was depleted from the media, a glycerol feed batch was carried out for an additional 26 h. We fed glycerol (500 g/L glycerol, 24 mL PTM4 salt solution) at a flow rate calculated for a growth rate of 0.02 h-1 to ensure carbon limitation since it was shown that high glycerol concentration has a negative impact on the expression of Pgap[17]. Samples were taken and processed as described above for HPLC analysis of the PG content. In the later stage of the fermentation (after 42 h), PG concentration was high enough in the broth that no concentration step was required for detection and quantification.
2.6. Pichia-PG1-1 Growth on Crude-Glycerol Media
- A preliminary study on growth and production in MGY containing crude glycerol from a biodiesel production unit (Point Hope Energy, Midland, MI) instead of ACS grade glycerol was conducted. Crude glycerol was processed by a single step filter-sterilization prior to use and mixed with the other components of the minimum media (MM). First, we tested the Pichia-wt with different concentrations of crude glycerol (0 to 80 g/L) in the minimum media. Next, we grew Pichia-PG1-1 cell line with crude glycerol at 40 g/L and 24 h (220 rpm, 30℃) and assessed the PG production by HPLC.
3. Results
3.1. Design of the PG Metabolic Pathway Cassette
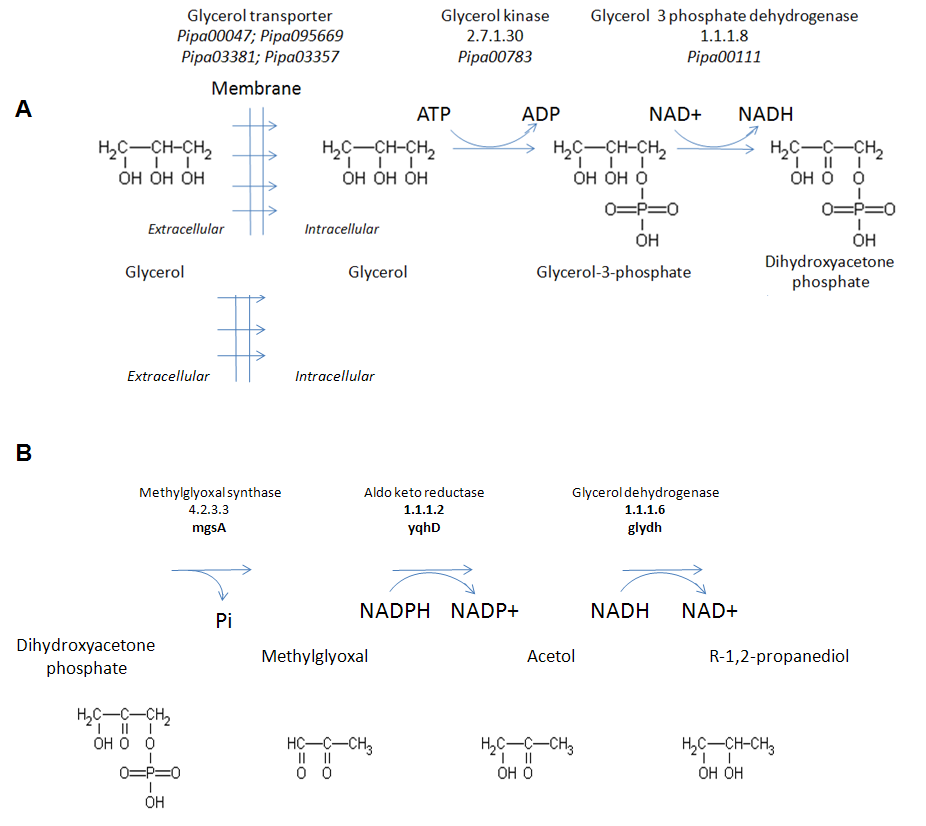
- The biosynthetic pathway we engineered in P. pastoris for production of PG from crude biodiesel glycerol is presented in Figure 2B. The pathway consisted of the following genes: mgsA gene from E. coli to produce the enzyme MG synthase[18], the yqhD gene from E. coli to produce the enzyme aldo-keto reductase[19], and the glydh gene from P. ofunaensis coding for glycerol dehydrogenase with acetol reductase activity[16]. When the 3 gene cassettes were combined, the construct was synthesized as a continuous sequence and cloned in pGAPZ (Fig. 3B; Table 1).
3.2. Pichia Transformation and Screening
- Transformed cells were grown at different level of Zeocin concentration and tested by PCR for specific gene integration of the PG biosynthetic pathway genes. Thirty clones out of the 60 we screened contained the PG biosynthetic pathway cassette. Fifteen clones were further processed.
|
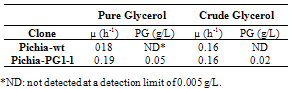
3.3. PG Determination in Culture Spent Broth and Enzyme Activities
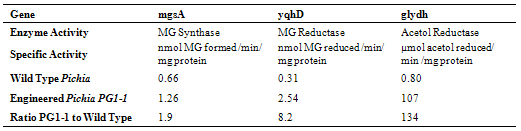
- Pichia clones containing the full PG biosynthetic gene were tested for PG production in shake flask cultures. A Pichia-wt was also grown for control. PG was detected by HPLC in concentrated spent broth of all the Zeocin®- resistant Pichia cell lines, and no PG was detected in the Pichia-wt concentrated spent broth. Thus, the biosynthetic pathway was successfully implemented in P. pastoris. In our experimental conditions, the detection limit for PG was determined to be at around 0.005 g/L. The cell line with the highest PG production (0.04 g/L) was used for further experiment and designated Pichia-PG1-1. Crude cell extracts were also tested by HPLC but no PG was detected (results not shown). Specific activities of the enzymes of the PG biosynthetic pathway were determined on extracts of Pichia-PG1-1 and compared to extracts of Pichia-wt. The activities of the PG biosynthetic enzymes were higher in the Pichia-PG1 than in Pichia-wt (see Table 2): about 134 times for the acetol reductase activity carried by Glydh, roughly 8 times for the MG reductase activity carried by YqhD, and only 2 times for the MG synthase activity carried by MgsA.
3.4. Pichia-PG1 Fermentation
- Fed-batch fermentation was run in 1.25 L fermentation vessel for 50 h. PG was detected in the fermentation broth, and the extracellular concentration increased steadily during the fermentation (Fig.5). PG concentration increased proportionally to yeast cell mass at an average of 0.2 mg PG/g WCW +/- 0.01. This result is expected since the promoter Pgap is a constitutive promoter and PG is stable in the fermentation broth. The highest PG concentration was 0.11 g/L at 49.5 hours. Specific PG productivity increased at the end of the glycerol batch, and decreased during the feed batch phase (Fig.5).
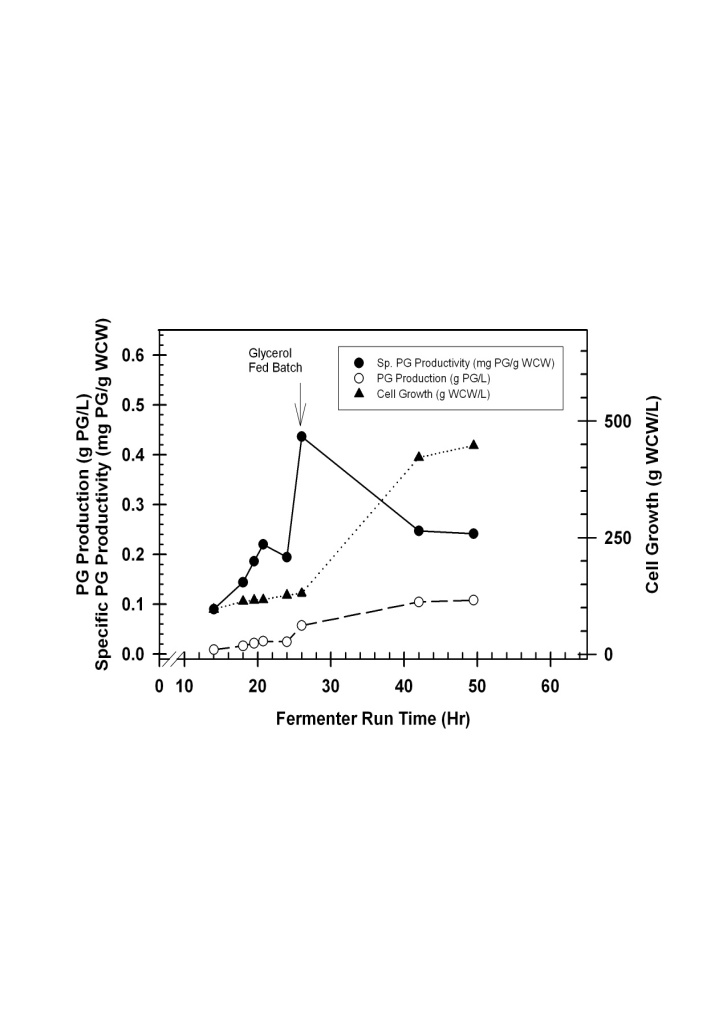 | Figure 5. Time course analysis of Pichia-PG1-1 cell line grown in the fermenter. The initial part of the batch glycerol phase is omitted for clarity |
3.5. Pichia-PG1-1 Growth on Crude-Glycerol Media
- Like previously observed[12], Pichia-wt grew on a minimum media (MM) containing up to 60 g/L of the crude glycerol, but grew slower at higher concentrations. The presence of inhibitory substances in the crude glycerol might be responsible for this effect. We then tested the impact of a 40 g/L crude glycerol MM on the Pichia-PG1-1 cell line and matched it against a Pichia-wt (Table 3). Growth parameters were similar for both cell lines: final biomasses were lower on the glycerol MM. No PG was detected in the Pichia-wt cultures. PG production of Pichia- PG1-1 was lower in glycerol MM than in MGY, which could be explained by the lower biomass obtained since production and growth are linked.
|
4. Discussion
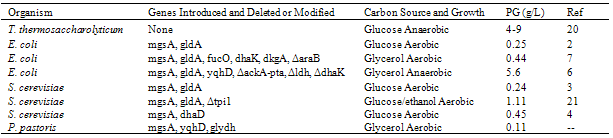
- In this proof of concept, we demonstrated that an engineered P. pastoris cell line aerobically biosynthesized PG from glycerol. The theoretical biochemical equation for the conversion is (Fig. 2):1 Glycerol + 1 ATP + 1 NADPH à 1 Propylene Glycol + 1 ADP + 1 Pi + 1 NADP+Thus, all the carbons of glycerol are conserved in the PG product, which is in contrast to ethanol production from glycerol where one carbon is lost as CO2. The theoretical biochemical equation for production of PG from glucose is[20]:1 Glucose à 1.5 PG + 1.5 CO2This corresponds to the more reduced state of glycerol compared to glucose. In theory, when 1 g of glycerol is consumed, 0.83 g PG would be produced. However, in reality some glycerol must be consumed to produce ATP and NAD(P)H and maintain cell growth. For glucose, the theory predicts 0.63 g PG per g glucose, but the actual yields are much lower.To engineer the PG pathway, we inserted contiguously at the GAP locus in P. pastoris Y11430 wild type three engineered genes for aerobic PG production from glycerol. Two of the genes were from E. coli: mgsA for the enzyme MG synthase and yqhD for the enzyme MG reductase. The other gene was P. ofunaensis glydh for a glycerol dehydrogenase to catalyze the final step from acetol to PG (Fig. 2B). The coding sequence for each of the genes was optimized with Pichia codons for highly expressed genes. We used the strong, constitutive promoter Pgap for control of expression of each of the genes (Fig. 3). After establishing by gene specific PCR of genomic DNA from the transformants that each of the genes was present in the engineered Pichia cells, we selected a PG producing cell line (Pichia-PG1-1) using shake flask cultures and demonstrated that the 3 enzymes were active in Pichia cell extracts (Table 2). Next we grew the PG1-1 cell line aerobically in the fermenter on glycerol where the PG level reached 0.11 g/L and specific PG productivity peaked at 0.4 mg per g WCW (Fig.5). Based on these results, the PG biosynthetic pathway was successfully implemented and functional in engineered Pichia cells. We also demonstrated that PG was produced when waste glycerol from a biodiesel production facility treated by a simple filtration step was used as carbon source to grow aerobically Pichia cell line PG1-1 (Table 3). PG accumulated in the spent cell broth, so this small hydrophilic molecule is exported from the cells by endogenous membrane transporters or simple diffusion. For comparison to our results, we have summarized all the results for PG production in the various engineered organisms studied to date (Table 4). The highest titers of PG are produced in E. coli strains, which must be grown anaerobically especially when glycerol is the carbon source. By very extensive modification of E. coli metabolic pathways for fermentation products and gene augmentations, a PG level of 5.6 g/L with a yield of 213 mg PG per g glycerol consumed was achieved[6]. However, when grown in the fermenter, this strain of E. coli also produced: ~330 mg ethanol/g glycerol; ~150 mg formate/g glycerol; ~50 mg succinate/g glycerol; and ~50 mg acetate/g glycerol. Clearly, this high level of PG production is unique, but it is in a complex mixture of products listed above as well as glycerol that was not consumed at the end of the fermentation. For yeast, the levels of PG produced are much lower in the range of a few hundreds of mg of PG/L of culture. Thus, the results presented here are comparable to those from other yeast systems; especially in consideration of the simple manipulations that have been applied in the engineered Pichia stain.Thus, Pichia pastoris is a good platform to produce PG aerobically from crude glycerol. But in order to have a competitive and attractive industrial strain able to create value from biodiesel waste crude glycerol, the titer, productivity and yield of the Pichia glycerol to PG process must be increased. In evaluating the limitations on production of PG by Pichia cell line PG1-1, we assayed cell extracts for enzyme activities of the 3 genes we integrated into the Pichia genome (Table 2). For the 1st enzyme in the engineered PG pathway, methylglyoxal synthase, the specific activity was about 2 fold higher in Pichia-PG1-1 than Pichia-wt. For the 2nd enzyme, methylglyoxal reductase encoded by E. coli yqhD, the specific activity was about 8 fold higher in Pichia-PG1-1 than Pichia-wt. For the last enzyme, acetol reductase, the specific activity was 134 fold higher in Pichia-PG1-1 than Pichia-wt. This would make it appear that production of methylglyoxal is a factor limiting flux into the engineered PG pathway. However, we found pyruvate/lactate was produced along with PG in the spent broth of the Pichia-PG1-1 cell line (data not shown), which most likely results from the detoxification of excess methylglyoxal by action of methylglyoxalase I.Thus, it appears that the reduction of methylglyoxal to acetol by the action of YqhD is insufficient to keep up with the flux of this intermediate. Therefore, YqhD appears to be the bottleneck in the biosynthetic PG pathway especially since the last enzyme, acetol reductase, is more than 1000 fold more active than YqhD (Table 2). This suggests that increasing number of copies of the yqhD gene would enhance activity of the 2nd enzyme in the PG biosynthetic pathway, and perhaps increase the titer of PG. Nevertheless, to increase the gene copy number, more than one loci of insertion must be available. In the P. pastoris system, genes are inserted in the genome by single crossover between the transforming DNA and homologous sequence in the Pichia genome[11]. We used the pGAPZ plasmid as a backbone for our work, so the targeted insertion is driven by homologous sequences located in front of the Pgap and the corresponding DNA at the GAP locus. Multiple insertion events at one locus can happen, but it is a random event occurring at a low frequency[11]. Finding a multiple insertion clone necessitates time consuming and laborious screening. Moreover, it can lead to an uncontrolled genomic rearrangement, which can end up in gene loss and genomic instability. Therefore, we have started the development of a set of plasmids with different insertion loci to increment rationally the copy number of the 3 genes encoding PG synthetic pathway enzymes. Since industrial strains need to be antibiotic resistance gene free, we are implementing these plasmids with a removable Zeocin® cassette based on the LoxP/Cre technology[22].
|
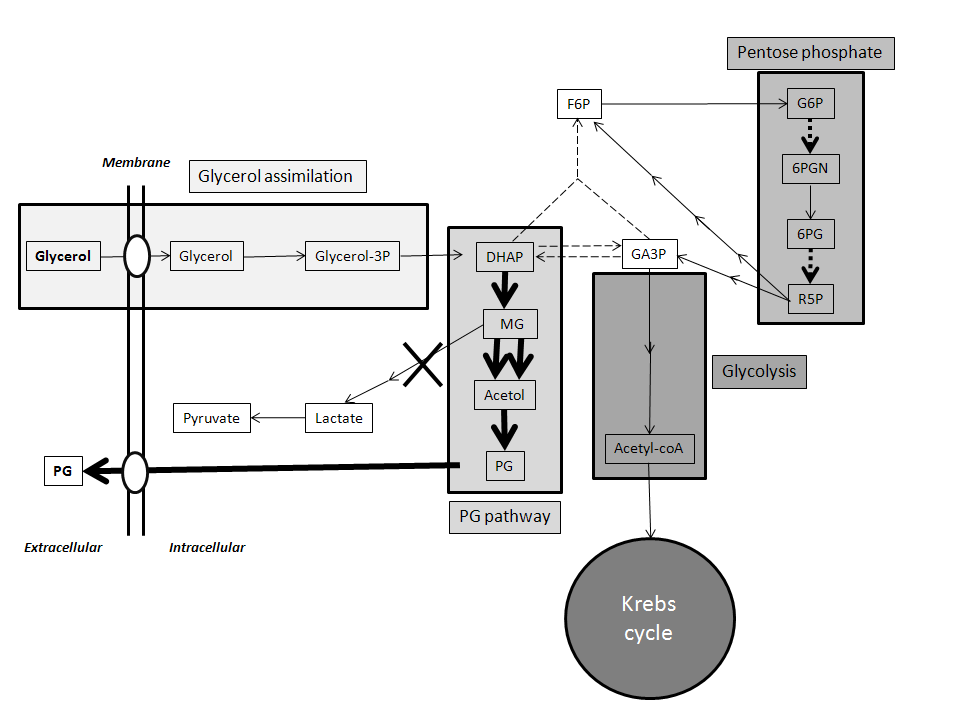 | Figure 6. Summary schematic of key pathways in P. pastoris and proposed modifications to increase PG production. Glycerol is assimilated to DHAP and then converted to PG via the metabolically engineered pathway, shown with bold arrows. The 2nd enzyme in the pathway will be increased by augmenting copy number of the yqhD gene, shown by double bold arrows. Conversion of MG to lactate and pyruvate catalyzed by methylglyoxalase 1, which drains carbon away from PG production, will be blocked by knocking-out the gene for this enzyme, shown by bold X. Additional enhancements of PG production may be achieved by attenuating expression of triose phosphate isomerase, shown by dashed line; and by increasing the expression levels of enzymes in the pentose phosphate pathway for augmenting the cellular reducing power, dashed bold arrows |
ACKNOWLEDGEMENTS
- This study is based upon work supported by the Small Business Innovation Research program of the National Science Foundation, under Grant No. 1013619. The Michigan Emerging Technologies Fund, a program administered by the Michigan Small Business & Development Center in cooperation with the Michigan Economic Development Corporation, provided additional support. We would also like to thank Richard Shull, P.E., President of Point Hope Energy, Inc., Midland, MI, for providing biodiesel waste glycerol.
References
| [1] | Nilles, D., 2006. Combating the Glycerol Glut. Biodiesel magazine. http://www.biodieselmagazine.com/articles/1123/combating-the-glycerin-glut/, last accessed 20June2011 |
| [2] | Altaras, N.E., Cameron, D.C., 1999. Metabolic Engineering of a 1,2-Propanediol Pathway in Escherichia coli. Appl. Environ. Microbiol. 65 (3), 1180–1185 |
| [3] | Lee, W., DaSilva, N.A., 2006. Application of sequential integration for metabolic engineering of 1,2-propanediol production in yeast. Metab. Eng. 8, 58-65 |
| [4] | Jeon, E., Lee, S., Kim, D., Yoon, H., Oh, M., Park, C., Lee J., 2009. Development of a Saccharomyces cerevisiae strain for the production of 1,2-propanediol by gene manipulation. Enz. Microbiol. Tech. 45 (1), 42-47 |
| [5] | Soucaille, P., Meynial-Salles, I., Voelker, F., Figge, R., 2008. Micro-organisms and methods for production of 1,2 propanediol and acetol. WO2008/116853 |
| [6] | Clomburg, J. M., Gonzalez, R., 2011. Metabolic engineering of Escherichia coli for the production of 1,2-propanediol from glycerol. Biotech. Bioeng. 108 (4), 867-879 |
| [7] | Mampel, J., Meurer, G., Eck, J., 2010. Production method. WO 2010/012604 Al |
| [8] | Barbier, G.G., Joshi R., Campbell, E.R., Campbell, W.H., 2004. Purification and biochemical characterization of simplified eukaryotic nitrate reductase expressed in Pichia pastoris. Prot. Express. Purif., 37, 61-71 |
| [9] | Campbell, W.H., Barbier, G.G., Campbell, E.R. 2007. Simplified eukaryotic nitrate reductase. US7262038 |
| [10] | Campbell, W.H., Song, P., Barbier, G.G., 2006. Nitrate Reductase for Nitrate Analysis in Water. Env. Chem. Let. 4, 69-73 |
| [11] | Cregg, J.M., Pichia protocols, 2007, 2nd ed., Volume 389, Humana press, Totowa, NJ |
| [12] | Tang, S., Boehme, L., Lam, H., Zhang, Z., 2009. Pichia pastoris fermentation for phytase production using crude glycerol from biodiesel production as the sole carbon source. Biochem.Eng. J. 43 (2), 157-162 |
| [13] | Sola, A., Maaheimo, H., Ylonen, K., Ferrer, P., Szyperski, T., 2004. Amino acid biosynthesis and metabolic flux profiling of Pichia pastoris. Eur. J. Biochem. 271, 2462–2470 |
| [14] | De Schutter, K., Lin, Y.-C., Tiels, P., Van Hecke, A., Glinka, S., Weber-Lehmann, J.,Rouzé, P., Van de Peer, Y., Callewaert, L., 2009. Genome sequence of the recombinant protein production host Pichia pastoris, a methylotrophic yeast. Nature Biotech. 27, 561-566 |
| [15] | Mattanovich, D., Graf, A., Stadlmann, J., Dragosits, M., Redl, A., Maurer, M., Kleinheinz, M., Sauer, M., Altmann, F., Gasser, B., 2009. Genome, secretome and glucose transport highlight unique features of the protein production host Pichia pastoris. Microb. Cell Fact. 2, 8-29 |
| [16] | Yamada-Onodera, K., Nakajima, A., Tani, Y., 2006. Purification, characterization, and gene cloning of glycerol dehydrogenase from Hansenula ofunaensis, and its expression for production of optically active diol. J. Biosci. Bioeng. 102 (6), 545-551 |
| [17] | Waterham, H.R., Digan, M.E., Koutz, P.J., Lair, S.V., Cregg, J.M., 1997. Isolation of the Pichia pastoris glyceraldehyde-3-phosphate dehydrogenase gene and regulation and use of its promoter. Gene 20 (186), 37-44 |
| [18] | Saadat, D., Harrison, D.H., 1999. The crystal structure of methylglyoxal synthase from Escherichia coli. Structure Fold. Des. 7 (3), 309-317 |
| [19] | Ko, J., Kim, I., Yoo, S., Min, B., Kim, K., Park, C., 2005. Conversion of methylglyoxal to acetol by Escherichia coli aldo-keto reductases. J. Bacteriol. 187 (16), 5782-5789 |
| [20] | Cameron, D.C., Altaras, N.E., Hoffman, M.L., Shaw, A.J., 1998. Metabolic engineering of 1,2-propanediol pathway. Biotechnol. Prog. 14, 116-125 |
| [21] | Jung, J.-Y., Choi, E.S., Oh, M.K., 2008. Enhanced Production of 1,2-Propanediol by tpi1 Deletion in Saccharomyces cerevisiae. J. Microbiol. Biotechnol. 18 (11), 1797–1802 |
| [22] | Marx, H., Mattanovich, D., Sauer, M., 2008. Overexpression of the riboflavin biosynthetic pathway in Pichia pastoris. Microb. Cell Fact. 29, 7-23 |
| [23] | Nguyen, H. T. T., Nevoigt, E., 2009. Engineering of Saccharomyces cerevisiae for the production of dihydroxyacetone (DHA) from sugars: A proof of concept. Metabol. Eng. 11, 335-346 |
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-Text HTML
Full-Text HTML