-
Paper Information
- Next Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Genetic Engineering
p-ISSN: 2167-7239 e-ISSN: 2167-7220
2011; 1(1): 1-5
doi: 10.5923/j.ijge.20110101.01
Cytochrome C Oxidase Subunit-1(COX1) Gene in Tilapia (Oreochromis Niloticus): its Cloning and Characterization
Iman M. K. Abumourad
Department of hydrobiology, National Research Center, Cairo, Egypt
Correspondence to: Iman M. K. Abumourad , Department of hydrobiology, National Research Center, Cairo, Egypt.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
The current study aied to identify cytochrome c oxidase subunit 1 (ONCOX1) in Tilapia (Oreochromis niloticus) immunized by formalin-killed Flavobacterium columnarae. Suppressive subtractive hybridization (SSH) was utilized to construct a cDNA library and a semi-quantitative RT-PCR analysis used to examine Oreochromis niloticus cytochrome c oxidase subunit 1 (COX1) gene expression. The complete sequence of the COX1 has been deposited in the GenBank Database and assigned the accession number (GE647880). COX1 cDNA is composed of 1139 bps with a 1107 bps open reading frame, the predicted gene product is 369 amino acid with molecular weight of 40.16 kDa. The amino acid sequence revealed high identity with subunit 1 from Oreochromis mossambicus. Compared to ß-actin, the semi-quantitative RT-PCR revealed that ONCOX1 expressed in tissues of stimulated fish as up-regulated gene suggesting that this member of COX genes is probably involved in the general immune response against the pathogenic bacteria.
Keywords: Tilapia, Cytochrome C Oxidase , Gene Expression, Protein Characterization
Cite this paper: Iman M. K. Abumourad , "Cytochrome C Oxidase Subunit-1(COX1) Gene in Tilapia (Oreochromis Niloticus): its Cloning and Characterization", International Journal of Genetic Engineering, Vol. 1 No. 1, 2011, pp. 1-5. doi: 10.5923/j.ijge.20110101.01.
1. Introduction
- Cytochrome c is a small, highly conserved, heme- containing bifunctional protein; it has been studied extensively, not only for its role in electron transport, but also for its role in apoptosis, it releases from the mitochondria in response to specific apoptotic stimuli (Wang X., 2001). Cytochrome c oxidase (COX) is an oligomeric enzymatic complex located in the inner membrane of mitochondria which is consists of three large primarily catalytic subunits encoded by the mitochondrial genome and 10 smaller subunits encoded by nuclear genes .The nuclear encoded subunits are thought to modulate enzyme function. In vertebrates, COX is composed of 13-subunits, and a further 30 proteins are required for proper COX assembly (Ettickan et al, 2004). In eukaryotes this enzyme complex is located in the mitochondrial inner membrane. It is considered to be a major site of regulation of mitochondrial oxidative phosphorylation. This rate-limiting enzyme is also implicated in the production of reactive oxygen species (ROS) under oxidative stress conditions (Lee et al, 2001, 2003 and Vijayasarthy et al., 2003). COX1 encodes an important enzyme involved in the oxidation phosphorylation pathway and thus energy production. In an attempt to characterize differential gene expression during cDNA library construction from tilapia immunized by F. columnarae, we could recognize ONCOX1 as up-regulated gene. The specific hypothesis for this study is that gene characterization and detectable changes in gene expression directed by certain pathobiological conditions may identify genetic information that are important for understanding the host-pathogen relationship and host immune responses., thus in this paper we summarize the identification of a cytochrome c oxidase subunit 1 gene utilizing available protein structure prediction methods based on the predicted protein sequence. The sequence we have identified and its predicted corresponding structure is the first functional annotation for this family of proteins in O.niloticus.
2. Material and Methods
- Tissue preparation and RNA extractionOreochromis niloticus 12 individuals were obtained from Wuhan, China. The fish were held in the laboratory, acclimatized in recirculating freshwater tank at room temperature under natural photoperiod for a week and fed with commercial pellets at a daily ration of 3% of their body weight. They were devided into two groups, the first were stimulated for four days each with intraperitonial injection of0.5 ml 107 formalin killed Flavobacterium columnarae, the other group were injected with PBS and used as a control. Two F.colmnarae injected tilapia, 24.5 cm and in fork length were dissected, gills, liver; spleen, head kidney and intestine were dissected out for the construction of the infected group subtracted cDNA library. One control (PBS injected fish) 23.7 cm in fork length was used also for the subtracted library with the mentioned organs dissected out. Total RNA was prepared for RT-PCR using Trizol Reagent (Invetrogen, USA). The mRNA was then isolated from total RNA using the poly Atract Isolation System (Promega). Concentration of mRNA was determined using a spectrophotometer.SMART cDNA synthesis and Suppressive subtraction hybridization (SSH)Complimentary DNA was synthesized andamplified using a CLONTECH cDNA Synthesis Kit by following the the manufacure's instruction. The mRNA obtained from tester and driver samples was subjected to SSH and selective PCR amplification using PCR-select cDNA subtraction kit to construct a cDNA library.Cloning and screening for differential expression by dot blotting and sequence analysissecondary PCRs products of stimulated and unstimulatd directly inserted into pGEM-T using a pGEM-T Easy Vector System ( Promega), which was then transformed into Escherichia coli and screened by the white colonies which were selected and amplified with nested PCR primer 1(5’-TCGAGCGGCCGCCCGGGCAGGT-3’) and nested PCR primer 2(5’- AGGGTGGTCGCGGCCGAGGT). 3 µl of PCR product were denatured in 3 ml of NaOH. Two identical H-bond N+ nylon membranes were prepared by loading 1 µl of denatured PCR product of each clone on the same location, after 5 min. neutralization in Tris-Hcl (pH7.5), the membranes were baked for 30 min at to cross link the cDNAs.Forward-subtracted cDNAs were digested with Rsa1 and labeled as probes with digoxigenin using a DIG High Prime system (Boehringer Mannheim) by following the manufacture’s instruction. Positive clones were sequenced using dedoxy chain sequencer (ABI Applied Biosystems Model 337).Sequence and phylogenetic analysisSequences were compared with the sequences in the database using the BLASTX program at the web server of the National biotechnology information. Protein prediction was performed using software at the ExPASY Molecular Biology Sever (http:∕∕expasy.pku.edu.cn) and SAPS program (Statistical Analysis of Protein Sequences). Sequences were aligned, employing the distance matrix; a distance-joining tree was constructed and a Phylogenetic analysis was carried out for the deduced amino acid sequences of cytochrome c oxidase subunit 1(COX1) using Clustal W version 1.83(Thompson et al. 1994), Sequence database alignments and comparisons were done with the BLAST family of programs (blastx,) against database specifications of non-redundant protein, SWISS-PROT which were available from the BLAST website at the National Center for Biotechnology Information webserver, (http://www.ncbi.nlm. nih. gov/ blast/) (Altschul et al. 1997). A signal peptide was predicted using Signal P 3.0 and analysis for transmembrane helices was applied using SOSUI dumbbell server.Semi - quantitative RT - PCR for tissue expression of COX1Total RNA from different mixed tissues was treated with DNase, 2 µg RNA was reverse transcribed with M-MLV reverse transcriptase using hexanucleotides (Promega) to prim the reaction. The first strand cDNA was used as templates for RT-PCR with a pair of COX1 specific primers designed, forward 5’- TTGGACACCCTGAAGT-3’ and reverse: 5’ GTGGCGGAAGTAAAGT3’.The PCR cycling parameters were one cycle of 94℃ for 5 min, 35 cycles of 94℃ for 30 s, 48℃ for 30 s and 72℃ for 1 min, with a final extenssion step of 72℃ for 10 min. The RT-PCR products were analyzed by electrophoresis on 2% agarose gel with PCR products derived from beta actin of the infected and non infected tilapia as controls, documented with documentation system. Semi-quantitative assessment of mRNA levels were determined by quantifying the band's intensity of PCR products using GeneTools in the gene expression relative to beta actin were tested between infected and uninfected fish using t-test.
3. Results
- Isolation of differentially expressed clonesThe comparison of pooled RNA samples from Oreochromis niloticus immunized by Flavobacterium columnarae against unimmunized healthy Oreochromis niloticus yielded multiple differentially expressed clones; some were suggested as up-regulated genes in the process of immunization as revealed by dot blot analysis and RT-PCR.Characterization of cDNA sequence of ONCOX1The complete sequence of the COX1 has been deposited in the GenBank Database (http://www.ncbi.nlm.nih.gov/ Genbank/) and was assigned the GenBank nucleotide accession number (GE647880). Blast queries of the sequenced nucleotides of COX1 demonstrated that the deduced amino acid sequence contains 1139 nucleotide bps with 1107 bps open reading frame coding for a protein of 369 aa and molecular weight 40.16 kda (Fig. 1).
 | Figure 1. Deduced amino acid sequences of ONCOX1 |
4. Discussion
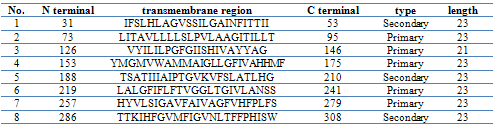
- Suppression subtractive hybridization (SSH) has led to the enrichment of specific expression genes as the non-specific expression of beta actin was reduced to about 215 time lower after subtraction, suggesting that non-specific expressed genes have been avoided and immunization- specific genes have been enriched efficiently in this study. The identification of new genes involved in the bacterial immunization provides the foundation for further research on the immunoilogical interaction between host and the bacteria, In this study, cDNA library construction in case of O. niloticus immunized by F. columnarae introduced some new immune genes, one of them was ONCOX1.dynamic change in an environment, disease hazards and the host-pathogen relationship provided signals for identifying differentially expressed genes which may play an important role in the immune system responses. During construction of cDNA library from O. niloticus immunized by F. columnarae, we could identify and characterize cytochrome c oxidase subunit 1 gene as up-regulated gene.ONCOX1 sequence was found to be homologous to other known cytochrome c oxidases with varying levels of sequence identity and high similarity with Oreochromis mossambicus (accession no.YP_272017) and also to human (accession no.BAA07292) and several vertebrates COX1, it appears to have structural features similar to the largest subunit of the heme/copper-requiring cytochrome c.The prediction of O-GalNAc (mucin-type) for glycosylation sites in ONCOX1 proteins indicated the absence of any glycosylation sites. Multiple methods of transmembrane prediction were used to enable a consensus confirmation of predicted transmembrane helices. The TM region search for ONCOX1 by SOSUI analysis program beta version suggests that ONCOX1 is a membrane protein with 8 transmembrane helices(TM). Transmembrane helices were found in the positions 29-53,72-95, 126-145, 152-171, 192-211, 218-237, 258-282 and 289-306 as showed in table 1. The objective of this TM search step was to identify and map out regions of transmembrane helices to confirm our sequence database based hypothesis of the putative sequence being identified as a cytochrome c oxidase subunit I. Cythochrome c oxidases are known to have these transmembrane regions and the identification of these regions served as a confirmatory step in gauging the validity of the predicted structure (Mohamed et al., 2003). The observation from the transmembrane prediction step is consistent with the subunit I of most other haem copper oxidases and acts as confirmatory data for correctness of the protein fold generated by the analysis program.A final taxonomic system for the animal kingdom will probably include at least 10 million species partitioned among more than a million genera (Paul et al., 2003). Given such high diversity, there is a growing realization that it is critical to seek technological assistance for its initial description and its subsequent recognition (Godfray 2002; Blaxter 2003). Recent investigations have suggested the feasibility of creating identification systems reliant on the analysis of sequence diversity in small segments of DNA (Tautz et al. 2003). Hebert et al. (2003) focused this discussion by proposing that a DNA barcoding system for animal life could be based upon sequence diversity in cytochrome c
|
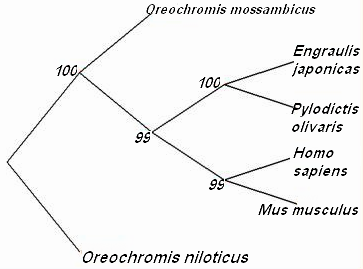 | Figure 3. phylogenetic distance tree showing the relationship among the COX1 from different vertebrates |
 | Figure 4. Tissue specific expression revealed by RT-PCR in Oreochromis niloticus; 1=βactin 2=immunized fish, 3= unimmunized fish |
5. Conclusions
- It could be concluded that killed F. columnarae did stimulate the immune response in the host fish, tilapia, some immune genes could be identified and characterized, one of these genes is COX1, which may have a role and included in the immune response to bacterial infection, thus the present study provides some information for further research on the structure of COX1 gene as an immune gene in relation to bacterial infection.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-Text HTML
Full-Text HTML