-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Ecosystem
p-ISSN: 2165-8889 e-ISSN: 2165-8919
2016; 6(2): 35-42
doi:10.5923/j.ije.20160602.02

Root Flavonoids of Convolvulus L. Species in Markazi Province, Iran
Batoul Bahrami1, Mitra Noori2, Amir Mousavi3, Ahmad Khalighi1, Aliashraf Jafari4
1Department of Horticulture, Islamic Azad University, Science and Research Branch, Tehran, Iran
2Department of Biology, Faculty of Science, Arak University, Arak, Iran
3Department of Plant Biotechnology, National Institute of Genetic Engineering and Biotechnology, Tehran, Iran
4Department of Rangeland, Research Institute of Forests and Rangelands, Tehran, Iran
Correspondence to: Mitra Noori, Department of Biology, Faculty of Science, Arak University, Arak, Iran.
| Email: |  |
Copyright © 2016 Scientific & Academic Publishing. All Rights Reserved.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

In order to study the root flavonoids pattern of Convolvulus roots in 11 populations from four species (C. arvensis, C. commutatus, C. lineatus and C. pilosellaefolius) originated from Markazi Province, Iran, two dimensional paper chromatography (2-DPC) and thin layer chromatography (TLC) were used. Results indicated that the roots contained flavonoid sulfates and flavones C and C-/O glycosides, apigenin, chrysin, genistein, kaempferol, luteolin, rhamnetin, rutin, and vitexin. hesperidin, isorhamnetin, morin, quercetin and tricine was not found in any of the taxa. Vitexin was the most found flavonoids in root. There was not any aglycone in the studied populations. All of populations had naringenin with the exception of CBB5, CBB2 and CBB6 populations. Myricetin was found in all of populations with the exception of CBB9 (C. commutatus). Populations of CBB19 (C. arvensis) had the most number of total flavonoids and C. lineatus and CBB9 (C. commutatus) had the least one.
Keywords: Convolvulus, Root, Chemotaxonomy, Chromatography, Flavonoid, Iran
Cite this paper: Batoul Bahrami, Mitra Noori, Amir Mousavi, Ahmad Khalighi, Aliashraf Jafari, Root Flavonoids of Convolvulus L. Species in Markazi Province, Iran, International Journal of Ecosystem, Vol. 6 No. 2, 2016, pp. 35-42. doi: 10.5923/j.ije.20160602.02.
Article Outline
1. Introduction
- Flavonoids are found in fruits, vegetables, grains, bark, roots, stems, leaves and flowers [1, 2]. Root flavonoids play significant roles in protecting the plants against pests and diseases, regulating root growth and functions, influencing different aspects of nitrogen cycle and exerting allelopathic growth effects. They also constitute an essential source of Pharmaceuticals [3]. The flavonoid pathway produces a diverse array of plant compounds with functions in UV protection, as antioxidants, pigments, auxin transport regulators, defense compounds against pathogens and during signaling in symbiosis [4]. Flavonoid synthesis and accumulation is often very specific for certain cell types. For example, along the length of a root, flavonoids are often accumulated at the root tip and in root cap cells. Specific flavonoid end-products are also localized to specific cell types where they could have functions in regulating [5, 6]. Within the cell, flavonoids also show specificity for their location. Flavonoids have been localized to the nucleus, the vacuole, the cell wall, cell membranes, and the cytoplasm [7-10]. To date, >10000 flavonoids have been identified in plants, and their synthesis appears to be ubiquitous in plants [11]. The flavonoid pathway is one of the best studied biosynthetic pathways of specialized metabolites. Flavonoids are phenylpropanoid metabolites, most of which are synthesized from p-coumaroyl-CoA and malonyl-CoA and share their precursors with the biosynthetic pathway for lignin biosynthesis [12]. However, some rare flavonoids are synthesized from CoA esters of substrates such as cinnamic acid or dihydro-coumaric acid [13]. Their diversity stems from the generation of a number of basal flavonoid structures that include flavones, flavonols, flavan-3-ols, flavanones, isoflavonoids, isoflavans, and pterocarpans. The flavonoid skeleton can be modified by glycosylation, malonylation, methylation, hydroxylation, acylation, prenylation, or polymerization, leading to the diversity of end-products [14]. These substitutions have important effects on flavonoid function, solubility, mobility, and degradation [4]. Flavonoids can also be transported within and between cells and tissues. Within the cell, flavonoids are likely to move via vesicle-mediated transport or through membrane-bound transporters of the ABC (ATP binding cassette) or MATE (multidrug and toxic extrusion compound) families [15]. Flavonoid transport into vacuoles can be achieved by conjugation of glutathione with flavonoids in the cytoplasm, followed by ATP-driven transport via glutathione S-transferasepumps [16, 17, 18]. Long-distance transport of flavonoids is less well understood but has been demonstrated in Arabidopsis, where application of flavonoids to the root or the shoot led to their transport towards distal tissues [19].Plant chemosystematics is the application of chemical data to systematic problems. It is a rapidly expanding interdisciplinary field concerned with using chemical constituents for explaining relationships between plants and inferring phylogeny [20]. Secondary metabolites especially flavonoids are valuable, widely and effectively used in chemosystematics [21]. Flavonoids occure widely in plants and are a biologically major and chemically divers group of secondary metabolites and are popular compounds for chemotaxonomic surveys of plant genera and families [22]. They have popular characters for chemosystematics studies because the almost universal presence of flavonoids in vascular plant, their structural diversity, the fact that each species usually contains several flavonoids and chemical stability of many flavonoids in dried plant material. Flavonoid profiles using different chromatographic techniques are easily obtained and are reasonably easy to identify using published UV spectra data and available standards. They often show correlations with existing classifications at these levels, and support revisions of existing classifications at the family, genus and species level [23]. Plant phenolic patterns appear to be more useful for studying relationships within relatively narrow taxonomic limits, e. g. at the species and genus level [22, 24, 25]. Today, flavonoids are used for making antitumoure, anticancer, antibacterial, antiviral, antifungal drugs and insecticides. There are some studies in this connection from mosses and liverworts [26], ferns [22], to the angiosperms [27, 22, 28, 29].The Convolvulaceae (Morning Glory family) is a beautiful family which is widely cultivated as ornamentals [30]. Convolvulus from Convolvuleae (Convolvulaceae) has about 250 species worldwide [31] and 60 species in Iran [32]. The family is widely distributed in cold regions, temperate, subtropical and tropical areas all over the world. There are several chemical studies both in the family Convolvulaceae and the genus Convolvulus. Alkaloids, flavonoids, coumarins, sterols, saponins, resin glycosides, tannins and stilbene derivatives have been isolated from plants of this genus [33, 34, 35]. Alkaloids were reported from Convolvulus [36], while acylated anthocyanins have been identified in many genera e.g. Ipomoea [37], Convolvulus [38] and Calystegia [39]. Rutin (quercetin 3-rutinoside), isoquercetin and kaempferol, 3- rhamnoglucoside and the coumarinsscopoletin and umbelliferone have been recorded in the family [38]. Rizk (1982) and El-Nasr (1982) studied Convolvulus species and showed the presence of flavonoids but they did not identify their constituents [8, 34]. Some species of Convolvulaceae produce resin where glycosides from which D-glucose, D-rhamnose, D-fucose and D-quinovose have been isolated [40]. Studies on phenolic constituents of Ipomoea eriocarpa and I. sindica using 2-D paper chromatography and TLC methods showed seven flavonoid glycosides in I. eriocarpa and four of these compounds in I. sindica [41]. A novel flavonoid, quercetin 7-methyl ether-3, 3'-disulfate, was obtained from the roots of Argyreia mollis (Convolvulaceae), together with the known compounds quercetin 7-methyl ether-3-sulfate and kaempferol 7-methyl ether-3-sulfate [42]. Menemen et al. (2002) studies on 20 Convolvulus taxa showed that aglycone pattern was useful for separation of some species in the genus [35]. Isorhamnetin 3-gucoside, quercetin 3-glucoside and 3-galactoside and luteolin 5-glucoside were identified in C. mazicum. Atta et al. (2007) isolated kaempferol and quercetin from C. fatmensis Ktz [43]. Mojab et al. (2003) showed presence of flavonoids in C. arvensis [44]. Kaur and Kalia (2012) reported 7-O-β-D-glucoside, 3-O-β-Dgalactorhamnoside, 7-O-rutinoside, 3-O-α-Lrhamnosyl, 3-O-α-L-rhamnoside, Kaempferol -3-O-β-D-glucoside and quercetin -3-O-α-L-rhamnoside in root, aerial parts and flower of C. arvensis [45]. Madhavan et al. (2008) carried out physicochemical analysis on C. microphyllus and Evolvulus alsinoides and identified their phenolic compounds, but not the type of them [46]. Bhowmik et al. (2012) showed the presence of alkaloids, glycosides, coumarins and flavonoids in C. pluricaulis. Moreover, Andrade et al. (2012) found kaempferol in C. pluricaulis [47, 48]. Preliminary phytochemical investigation of aqueous and methanolic extract of roots of Argyreia nervosa revealed the presence of alkaloids, glycosides, amino acid, tannins and flavonoids which were further confirmed by thin layer chromatography [49]. Studies of Bahrami et al (2016) on four Iranian Convolvolus species leaf flavonoids showed existing flavonoid sulfates and flavones C and C-/O glycosides, quercetin, kaempferol, isorhamnetin, myricetin, rhamnetin, rutin, apigenin, chrysin, luteolin, vitexin, genistein, hesperidin and naringenin in all of them. Morin just found in C. arvensis and C. lineatus species had aglyconeswhile others lacked [50].This study presents the root flavonoid patterns of 11 collected Convolvulus (C. arvensis, C. commutatus, C. lineatus and C. pilosellaefolius) populations from different parts of Markazi Province, Iran for understanding flavonoids role in Convolvulaceae chemotaxonomy. This is a novel report on Convolvulus root flavonoid patterns. In addition, some of flavonoid types in C. arvensis were identified for the first time.
2. Materials and Methods
2.1. Plant Collection and Preparation
- Mature fresh roots of 11 Convolvulus populations were collected from different parts of Markazi Province, Iran during 2013 spring as described in Table 1. Samples were identified using available references [51, 52, 53]. Voucher specimens of each sample were prepared for reference as herbarium vouchers. Samples were air dried for detection and identification of their flavonoids.
2.2. Plant Extracts Preparation
- For a comparative analysis of the flavonoids, small extracts of all the accessions were prepared by boiling 200 mg of powdered air dried root for 2 min in 5 ml of 70% EtOH. The mixture was cooled and left to extract for 24 h. The extract was then filtered, evaporated to dryness by rotary evaporation at 40°C, and taken up in 2 ml of 80% MeOH for analysis by 2-Dimensional Paper Chromatography (2-DPC) [54].
2.3. Two-Dimensional Paper Chromatography (2-DPC)
- For the detection of flavonoids, ca 20 μl of each of the small extracts was applied to the corner of a quarter sheet of Whatman No 1 chromatography paper as a concentrated spot (10 applications of 2μl). The chromatogram for each sample was developed in BAW (n-BuOH-AcOH-H2O = 4:1:5; V/V; upper layer), 1st direction, and AcOH (=15% aqueous acetic acid), 2nd direction, with rutin (quercetin 3-O-rutinoside) as a standard. After development, the chromatograms were viewed in long wave UV light (366 nm) and any dark absorbing and fluorescent spots were marked. Rf values in BAW and 15% AcOH were calculated.
2.4. Flavonoids Identification
- After obtaining sufficient amounts of purified flavonoids, as in the case of the flavonoids from root of the populations, they were identified by means of UV spectroscopy using shift reagents to investigate the substitution patterns of the flavonoids and by acid hydrolysis to identify the aglycone and sugar moieties. Cochromatography with standards was also performed where possible [54, 55]. Flavonoid standards available for comparison during the study were apigenin, chrysin, genistein, hesperidin, isorhamnetin, kaempferol, luteolin, morin, myricetin, naringenin, quercetin, rhamnetin, rutin, tricine and vitexin (all obtained commercially from Merck, apigenin, luteolin and hesperidin from Sigma and the rest from Fluka).
2.5. Acid Hydrolysis and Identification of Flavonoid Aglycones
- A small amount of each purified flavonoid (ca 0.5 m was dissolved in 0.5 ml of 80% MeOH in a test tube. To this sample 2 ml of 2M HCl was added and the mixture was heated in a water bath at 100°C for 0.5 h. The solution was cooled; 2 ml of EtOAc was added and thoroughly mixed with the aqueous layer using a whirley mixer. The upper EtOAc layer was removed with a pipette, evaporated to dryness, dissolved in 0.5 ml of MeOH and applied as spots on thin layer chromatograms (cellulose). The TLC plates were run in three solvents alongside standards to identify the aglycone moiety [56].
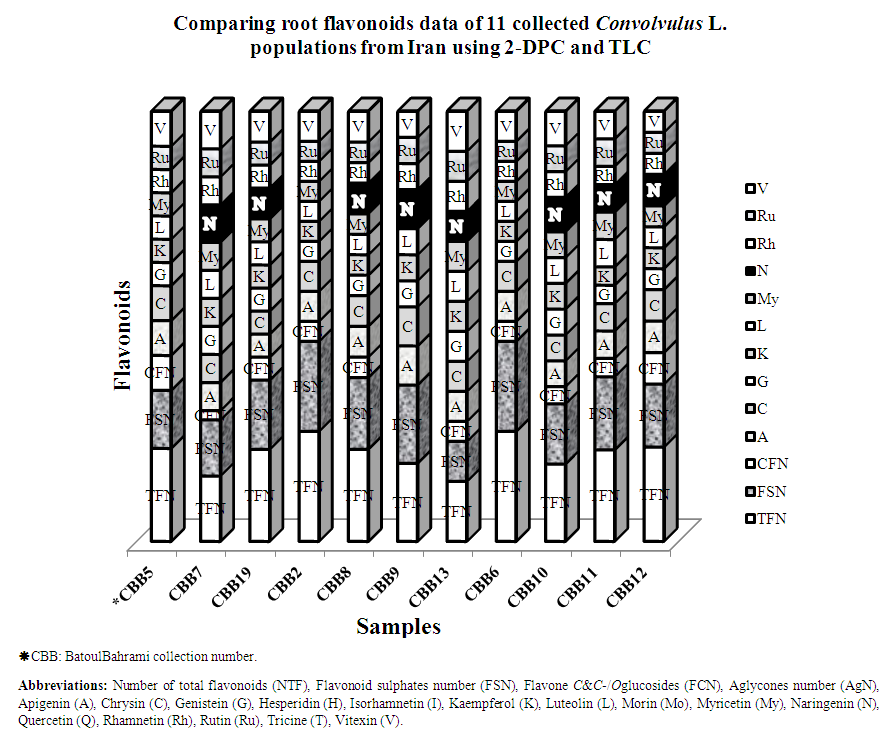
3. Results
- Results showed all of studied Convolvulus samples contained flavonoid compounds in their roots. Data in Tables 1 and 2 show the collection information and also 2-dimentional paper and thin layer chromatographical data of 11 studied convolvulus populations from Markazi Province, Iran. Figure 1 shows stacked column with a 3-D visual effect histogram for comparing root flavonoids data (number of total flavonoids, flavonoid sulphates number, flavone C-and C-/O-glucosides number and occurrence of apigenin, chrysin, genistein, kaempferol, luteolin, myricetin, naringenin, rhamnetin, rutin, and vitexin in the populations).As indicated in Table 1 and 2 and also Figure 1 hesperidin, isorhamnetin, morin, quercetin and tricine (syn: zwitterionic amino acid) as an isoflavone were not found in any of the taxa and vitexin was the most found flavonoids. Naringenin was found in all of the taxa with the exception of CBB5 (C. arvensis), CBB2 (C. commutatus) and CBB6 (C. pilosellaefolius). Also myricetin was found in all of the taxa with the exception of CBB9 (C. commutatus). CBB19 Population (C. arvensis) had the most number of total flavonoids, C. lineatus and CBB9 (C. commutatus) had the least one. Flavonoid aglycone was not detected in all taxa (Table 1 and Figure 1).
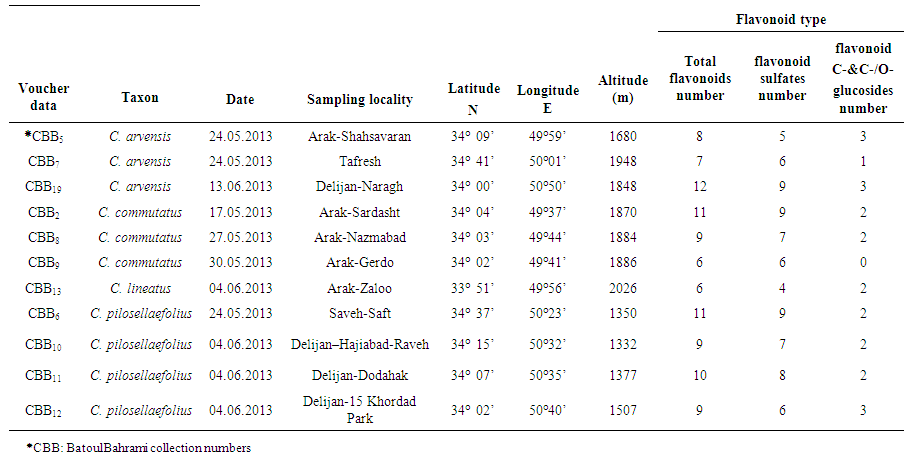 | Table 1. Collection information and root 2-Dimentional Paper Chromatography data for 11 studied Convolvulus populations from Markazi Province, Iran |
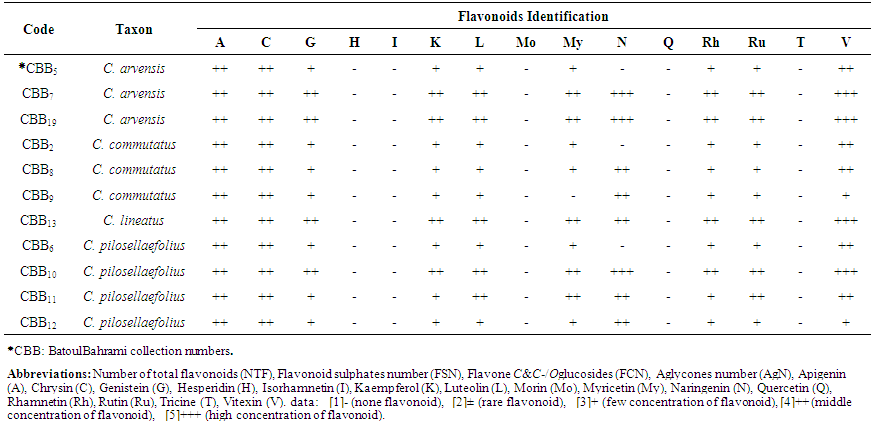 | Table 2. Thin Layer Chromatographical data of 11 studied Convolvulus populations’root flavonoid from Markazi Province, Iran |
4. Discussion, Conclusions and Recommendations
- Not only flavonoids are taxonomically important but are important as molecules of plant interactions with the environment. Flavonoids are molecules displaying various biological activities with relevance to plant physiology and development. Flavonoids not only participate in protection against harmful abiotic factors, but also allow for interactions with other plants and microorganisms. They are also emerging as regulatory and signaling molecules. In particular, their influence on plant development via interaction with an auxin transport network points toward their regulatory function in view of very low concentrations needed for the activity, indeed much lower than is necessary for effective radical quenching. Their presence in the nucleus points towards their role as transcriptional regulators [9, 10]. This function is already performed in regulation of microbial genes during mycorrhizal interactions. The effect of flavonoids on cellular signaling is well described in animal models where dietary flavonoids interact with many proteins of signaling cascades by directly binding to the ATP catalytic sites of protein kinases [57]. The possible interactions with MAP kinases in plants are still unexplored; however, it is likely that flavonoids may be responsible for mediating ROS-induced signaling cascades vital to cell growth and differentiation. It is evident that flavonoids allow plants to be an integrative part of their environment by responding to biotic and abiotic stimuli [58].Flavonoids can cause allelopathy. Allelopathy, the inhibition of plant growth and germination by other plants, plays an important role in parasitic and invading plants and can have far-reaching ecological consequences. In some cases, flavonoids have been implicated as allelochemicals in the rhizosphere [4]. Some flavones and flavonones that induce nod genes, such as luteolin and apigenin, have also been shown to evoke a strong chemoattractant response from the rhizobia, with different flavonoids attracting different Rhizobium species [59, 60]. Flavonoids have also been shown to regulate a number of other Rhizobium genes, including those for exo-polysaccharide synthesis, which is important for regulating defence responses in the host. For example, genistein at 1M concentration altered exo-polysaccharide concentration and composition in Rhizobium fredii cultures [61]. In addition, type III secretion systems, which play a role in nodulation in some rhizobia, as well as the production of exported proteins, can be induced by flavonoid exudates [62]. Proteome analysis also found a number of other proteins in response to host flavonoids, many of which await characterization [63]. Flavonoids and other phenolics have been found to inhibit a range of root pathogens and pests, ranging from bacteria to fungi and insects [64]. This has been attributed to their role as antimicrobial toxins [65] and anti- or pro-oxidants [66]. Their role within the plant as antioxidants is suspected to be protective, although clear evidence is lacking [67]. More than 4000-10000 varieties of flavonoids have been identified in different higher and lower plant species [11, 68]. The main flavonoid groups are flavones (e.g. luteolin), flavanone (e.g. naringenin), flavonols (e.g. kaempferol), anthocyanidins (e.g. pelargonidin) and chalcones (e.g. butein) [69]. In this research, the presence of three types of flavonoids including flavonols (kaempferol, myricetin, rhamnetin and rutin), flavones (apigenin, chrysin, luteolin, vitexin), isoflavones (genistein) and flavanone (naringenin) were reported in all of the studied species roots.There was not any flavone C-and C-/O-glucosides in population of CBB9 (C. commutatus) (Table 1 and Figure 1). It seems that the root flavonoids pattern would be not useful for delimitation of species and populations since the flavonoid was absent in one or more members of the taxon and the same flavonoid occured in an unrelated taxon as Harbone and Turner (1984) found in their researches [70]. Kaur and Kalia (2012) reported 7-O-β-D-glucoside, 3-O-β-Dgalactorhamnoside, 7-O-rutinoside, 3-O-α-L-rhamnosyl, 3-O-α-L-rhamnoside, kaempferol -3-O-β-D-glucoside and quercetin -3-O-α-L-rhamnoside in root, aerial parts and flower of C. Arvensis [54].Furthermore, Menemenet al. (2002) detected quercetin 3-mono- or di glycosides in all of 20 studied Convolvulus species, while kaempferol 3-mono- glycosides was just found in four taxa (C. arvensis, C. sabatius ssp. sabatius, C. sabatiussspmauritanicus and C. siculus ssp. elongatus). They reported presence of quercetin 3-mono- or di glycosides and cichorin in C. lineatus [35]. The flavonoid was found in addition to other flavonoid compounds in this species. Preliminary phytochemical investigation of aqueous and methanolic extract of roots of Argyreia nervosa revealed the presence of alkaloids, glycosides, amino acid, tannins and flavonoids [49]. Preliminary Phyto-chemical analysis and antioxidant activities of methanol extract of Argyreia roxburghii Choisy root that carry out by Baruah et al. (2014) indicated that the genus root is a potential source of natural antioxidant [71]. Analysis of extracts obtained from roots of Brassica alba revealed the presence of 20,30,40,50,60-pentahydroxy chalcone and 3,5,6,7,8-pentahydroxy flavone in their roots. Apigenin was also found in the roots [72], as we found it in this research. Noori (2014) investigated root flavonoids profiles from 10 populations of five Scirpus species from Iran for introducing chemotypes. Her results showed all of studied Scirpus species roots had vitexin, luteolin, rutin and rhamnetin [73].Although flavonoid compounds are taxonomically important and often show correlations with existing classifications at the family, genus, and species but rarely provide key characters since the flavonoid may be absent in one or more members of the taxon and the same flavonoid may occurs in an unrelated taxon [70]. These studies show that plant phenolic patterns appear to be more useful for studying relationships within relatively narrow taxonomic limits, e. g. at the species and genus level as Harborne (1994), Moor and Giannasi (1994), Noori et al. (2009) and Noori (2014) found in their works [22, 24, 25, 73, 74]. As we know that plant flavonoid pattern depends on genetics factors and ecological conditions and these parameters are effective on flavonoid production, it is believed that flavonoid patterns cannot always reveal the taxa differences.It is suggested that for more subtle results, studying other biosystematics characters would be required. In addition, molecular marker application along with the current research strategies could be useful and is recommended.
ACKNOWLEDGEMENTS
- The authors wish to thank of Arak University for its support throughout this project. Our special thanks go to Dr. Mozafarian, Mr. M. Golestani, Mr. M. Nezamdost and Mrs. H. Bahrami for generous assistance in collecting and determination of the studied samples.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML