-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Biological Engineering
p-ISSN: 2163-1875 e-ISSN: 2163-1883
2024; 8(1): 1-5
doi:10.5923/j.ijbe.20240801.01
Received: Oct. 27, 2024; Accepted: Nov. 22, 2024; Published: Dec. 11, 2024

The Role of CHEK2 Gene 1100delC rs555607708 Polymorphism in the Development of Breast Cancer in Uzbek Women
N. V. Khudoyberdiyeva1, 2, M. M. Abdullayeva1, Q. T. Boboyev3, M. N. Tillyashaykhov4, Sh. N. Ibragimov4, N. Sh. Avezov5
1PhD Researcher, National University of Uzbekistan
2Alfraganus University, Uzbekistan
3UzR SSV Republican Center of Specialized Hematology Scientific and Practical Medicine, Uzbekistan
4Republic Specialized Scientific and Practical Medical Center of Oncology and Radiology of the Republic of Uzbekistan
5Tashkent Pharmaceutical Institute, Uzbekistan
Correspondence to: N. V. Khudoyberdiyeva, PhD Researcher, National University of Uzbekistan.
| Email: |  |
Copyright © 2024 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

It is known that cervical cancer is the most common type of cancer among women and the most common cause of death of women with oncology. In addition to external and internal influences, genetic predisposition also plays a role in the origin of oncology. In addition to genes with high penetrance, such as BRCA1/2, which are responsible for the development of BC, genes with relatively low penetrance, such as ATM, CHEK2, BRIP1, PALB2, are currently being identified. It is known that the polymorphism frequencies of these genes have different indicators in different populations. Our investigations were focused on studying the prevalence of CHEK2 gene 1100delC rs555607708 polymorphism in the population of Uzbek women.
Keywords: CHEK2 gene, Breast cancer, 1100delC polymorphism, Oncology
Cite this paper: N. V. Khudoyberdiyeva, M. M. Abdullayeva, Q. T. Boboyev, M. N. Tillyashaykhov, Sh. N. Ibragimov, N. Sh. Avezov, The Role of CHEK2 Gene 1100delC rs555607708 Polymorphism in the Development of Breast Cancer in Uzbek Women, International Journal of Biological Engineering, Vol. 8 No. 1, 2024, pp. 1-5. doi: 10.5923/j.ijbe.20240801.01.
Article Outline
1. Introduction
- It is known that breast cancer is the most common and leading cause of death among women in recent years. Several factors are responsible for the origin of KBS, of which 5-10% are genetic predisposition [1]. Women with a strong history remain at high risk even in the absence of genetic mutations. The onset of BC is spontaneous in most cases, but mutations in certain genes have been implicated in the pathogenesis of BC, including ATM, BRAC1/2, BRAS2, TP53, and CHEK2 (checkpoint kinase2) in BC found to belong to the group of causative genes [2,3]. Cells are constantly exposed to endogenous and exogenous stresses, and even in the normal metabolic state, certain damages are observed in the DNA of 50,000-500,000 cells every day. This indicator increases even more under the influence of smoking tobacco products, various toxins, air pollution, nutritional chemical (artificial) additives. Single (SSB-single-strand breaks) or double (DSB-double) DNA breaks under the influence of free radicals of oxygen formed as a result of metabolism, ultraviolet rays (UV) or ionizing radiations (cosmic radiation, radiotherapy and medical procedures using X-rays) can cause chain breaks [4,5,6]. Although DSBs occur relatively rarely (~10 in one cell per day), they are considered very dangerous, and these changes cause chromosome rearrangements and loss of genetic material. All living organisms have developed complex but effective defense systems against SSB and DSB changes. This cascade of processes is called DDR (DNA Damage Response) and requires the participation of hundreds of proteins [7]. In order to maintain genetic integrity, a cascade of complex proteins works continuously and plays an important role in cell DNA repair (repair), cell cycle arrest or apoptosis. Like other proteins of the DDR, CHK2 controls the continuation of mitosis and meiosis, ensures genome stability of basal cells, and participates in response reactions when an organism is infected by CHK2 interaction with viral proteins and mitochondrial DNA damage [8]. Recent studies have shown that CHK2 pathogenic variants are associated with prostate, kidney, and thyroid cancers in addition to BC [9,10].The human kinase CHK2 was first identified in 1998 by Matsuoka et al. was identified by CHEK2 NM_0071194.4:c.1100del p. (Thr367fs) and later CHEK2 c. 1100del) variants were found [11]. In 2010, Stoltz et al. They found out that CHEK2 has an important property in mitosis. Also, CHEK2 is responsible for the correct and timely formation of the division nucleus during mitosis, the correct formation of kinetochores and the subsequent correct segregation of chromosomes [12]. CHEK2 is a gene suppressor that encodes the serine-threonine kinase CHK2 protein. This gene is located on the long arm of human chromosome 22 at position (locus) 12.1. CHK2 is a 65160 D protein consisting of 543 amino acids and has 3 main domains. 1. N-terminal cluster domain SQ/TQ (SCD)2. central domain (FNA)3. C-terminal domain (KD) of serine/threonine kinaseThe N-terminus, which is rich in serine-glutamine and threonine-glutamine pairs, mainly contains the phosphorylation sites of PI3K family kinases, including ATM and ATR. Between residues 112 and 175, there is a fork-shaped (FNA) domain, which is the center responsible for interaction with phosphorylated proteins. Residues 220-486 are located at the C-terminus, which is considered the canonical domain. FHA and SCD centers are common components of DDR proteins [13]. The catalytic function of CHEK2 is achieved by phosphorylation of a polypeptide center known as the T-loop (residues 366-406), which is located within the kinase domain, but away from the active site, like most kinases. Between residues 515-522 there is a signal center that directs the newly synthesized CHK2 protein to the subcellular component [7].
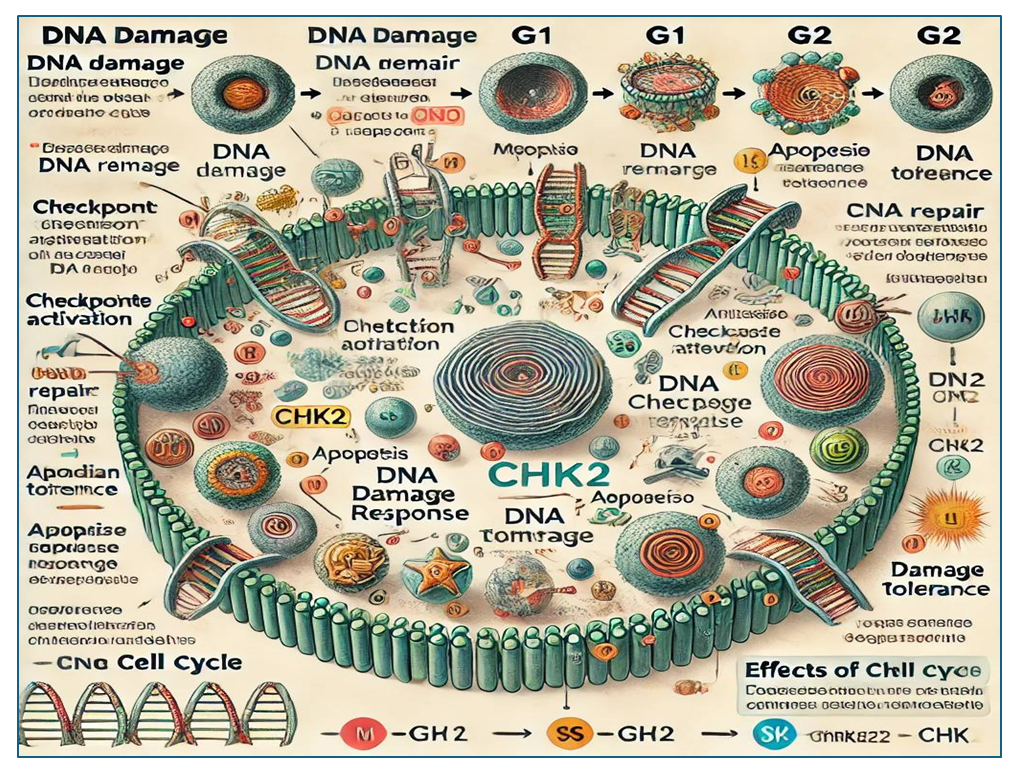 | Figure 1. CHK2 functions during DNA damage in human cells |
2. Research Materials and Methods
- For this study, 200 patients diagnosed with breast cancer based on the results of mammography and histology at the specialized Oncology and Radiology scientific-practical medical center of the Republic of Uzbekistan and it is Tashkent city branch mammology department and as a control group (control group) Blood samples were taken from 100 conditionally healthy Uzbek women. All molecular genetic tests were carried out in the Department of Molecular Medicine and Cell Technologies of the Republican Specialized Hematology Scientific and Applied Medical Center. DNA was isolated from the peripheral blood of patients and conditionally healthy Uzbek women using the "AmpliPrime Ribo-prep" kit (Next Bio, Russia). The quantity and quality of the extracted DNA was checked using a NanoDrop 2000 (Thermo Fisher Scientific, USA) spelterphotometer. The CHEK2 gene was tested for 1100delC (mutations mutation) according to the instructions of the Litex (Russia) genetic test kit. Polymerase chain reaction was carried out in the Corbett Research GRADIENT PALM CYCLER PCR Analyzers CG1-96 (Australia) amplifier, statistical analysis of the obtained results «WinPEPI 2016, Version 11.65 " and "EpiCalc 2000 Version 1.02" statistical computer programs were used.
3. Research Results
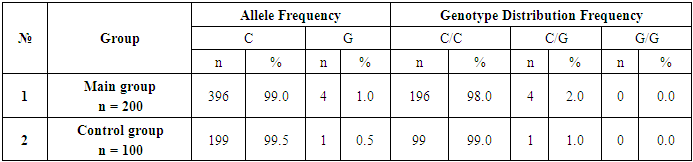
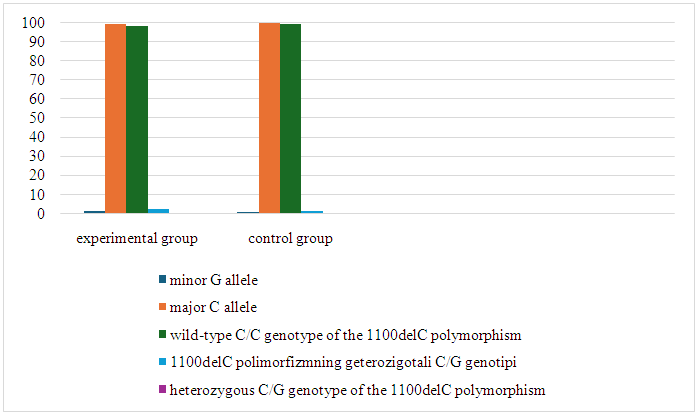
- A total of 300 Uzbek patients and conditionally healthy women were examined in the study. Among them, 200 patients aged between 30-65 years who underwent histological examinations, patients at different stages (T1-4, N0-3, M0/1) and 100 conditionally healthy patients aged 16-70 years on average. established women. The functionally dangerous allele G of CHEK2 gene 1100delC polymorphism was statistically more prevalent in breast cancer patients compared to healthy donors (1.0% and 0.5%, respectively); (χ2=0.4? p=0.5; OR=2.0; 95% CI: 0.22-18.11; RR=2.0; 95% CI: 0.22-17.77). The safe C allele, on the other hand, was more frequent in the control group than in the baseline group (99.5% and 99.0%, respectively, χ2=0.4? p=0.5; OR=0.5; 95% CI:0.05-4.48; RR=1.0; 95% CI:0.98-1.01). Thus, it was found that the dangerous G allele increases the probability and relative risk of breast cancer by 2 times. Also, CHEK2 gene 1100delC functionally natural C/C genotype was found in the highest frequencies - 99.0% in the control group and 98.0% in the group of patients with BC. However, the threshold of statistical differences reached a significant level: (χ2=0.4? p=0.5; OR=0.5; 95% CI: 0.05-4.48; RR=0.9; 95% CI: 0.96-1.02). The frequencies of the heterozygous C/G genotype of this 1100delC polymorphism were 2.0% (4/200) and 1.0% (1/100) in the primary and control groups. According to the results of the statistical analysis, the relative risk: RR=2.0; (95% CI: 0.22-17.66) and odds ratio: OR=2.0; (95% CI: 0.22-18.32). It is known that the C/G genotype increases the risk of developing breast cancer by 2 times. It should be noted that the rare homozygous genotype of the 1100delC polymorphism G/G was not detected in the studied patients and control groups.
|
|
4. Discussion
- It is known that genetic mutations in certain genes are one of the risk factors for breast cancer. In addition to genes with high penetrance, genes with moderate penetrance also exert their influence. The CHEK2 gene 1100delC rs555607708 polymorphism is one such gene [http://www.genecards.org/]. As far as we know, no investigations have been carried out to determine polymorphisms of the CHK2 protein in the population of Uzbekistan. The purpose of our work was to study the role of 1100delC rs555607708 polymorphism of CHEK2 in the origin of BC in Uzbek women. In order to determine whether mutations in the CHEK2 gene are related to the origin of BC, we performed genotypic analysis using standard PCR and Real-Time methods. In a Chinese population, patients with mutations in the CHEK2 gene were more likely to develop breast and/or ovarian cancer (23.1% vs. 8.6%, p=0.022) [15], while in a French-Canadian population, the frequency of the CHEK2 gene was 0% [16], It is 2.9% in France [17], 5.8% in Germany [18], 5.8% in Ashkenazi Jews, 8.9% in Great Britain, North America, and the Netherlands [19]. The frequency of 1100delC in women of Bashkir (Bashkortostan) was 0.9% [20], in Finland it was 12.2% [21], in Canada, the USA and Australia this indicator was 3.9% [22]. In the white-skinned aole population of South America, this indicator is 1% [23], in the Iron population it is 0.48% [24], in Poland it is 1.71% [25], in Brazil it is 4.9% [26], in Turkey and other Korea, Japan, India, the Philippines, Singapore, Malaysia, and Pakistan in the Asian region had very low percentages of CHK2 protein truncating mutations. [19,27]. Dutch population with 1100delC polymorphism mutation of CHEK2 protein is twice as likely to develop CHD compared to non-carriers of this mutation [16]. The frequency of 1100delC is 1.4% in the population of Northern European regions, including Finland, Great Britain and the Netherlands [22]. The frequency of 1100delC of CHEK2 in the Polish population was 0.25% [13]. From this study, it was found that the frequency of gene polymorphisms examined in the study corresponded to the frequency of Asian and Turkish peoples. As a result of our experiments, it became clear that the 1100delC polymorphism of the CHEK2 gene plays an important role in the development of breast cancer in Uzbek women. In particular, G allele of 1100delC polymorphism of CHEK2 gene and heterozygous C/G genotype are significant risk factors associated with increased probability of developing breast cancer (p>0.05). C allele and C/C homozygous genotype serve as reliable protective factors against the development of this pathology. Based on these results, we hypothesize that CHEK2 1100delC gene polymorphism can be used as a specific genetic marker to assess the risk of developing breast cancer.
5. Summary
- As we mentioned above, these tests were conducted for the first time in the Uzbek population. In the treatment of oncological diseases, it is very important to get a positive result, to save the patient's life, to treat it without harming the patient's health, and to make a timely diagnosis. We believe that the determination of the frequency of minor alleles is important for the selection of treatment methods, and it is important for patients to choose relatively radical surgical methods, and for timely prevention and prevention of the disease of relatives of patients with a mutation. According to the above results, we believe that the 1100delC polymorphism of the CHEK2 gene can be used as a specific genetic marker to assess the risk of developing BC.
6. Gratitude
- We express our gratitude to the employees of the mammology departments of the Specialized Oncology and Radiology Scientific and Applied Medical Center of the Republic of Uzbekistan and its Tashkent city branch, and the molecular medicine and cell technologies department of the Republic of Specialized Hematology Scientific and Applied Medical Center.
Abbreviations
- BC - breast cancerCHEK2 – checkpoint kinase2 DNA - deoxyribonucleic acidDDR - DNA Damage ResponseSSB - single-strand breaksDSB-double-strand breaksUV - ultraviolet rays
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML