-
Paper Information
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Biological Engineering
p-ISSN: 2163-1875 e-ISSN: 2163-1883
2012; 2(2): 18-22
doi: 10.5923/j.ijbe.20120202.03
Screening, Isolation and Biochemical Characterization of Novel Lipase Producing Bacteria from Soil Samples
Bhavani M 1, 2, G V Chowdary 3, David M 1, Archana G 1
1Department of Biotechnology, School of Engineering, Sir PadampatSinghania University, Udaipur, 313 601, India.
2Department of Biotechnology, AcharyaNagarjuna University, Guntur, 522 510, India.
3TRR College of Pharmacy, Pathancheru, Hyderabad, 502 301, India
Correspondence to: Bhavani M , Department of Biotechnology, School of Engineering, Sir PadampatSinghania University, Udaipur, 313 601, India..
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
Novel Lipase producing bacterial isolates were obtained from the soil samples collected from three distinct and distant locations in India. The soil samples were found to have the microbial content with a range of (2.1-2.4) x105 CFU/gm; with direct proportionality between the lipase producing isolates and the microbial content in the soil samples. Lipase producing isolates were screened by tributyrin agar plates with 0.1% Rhodamine B dye and further screened to obtain the best possible lipase producing isolate(s) subjected to variable temperature and incubation period. The isolate B1 produced the lipase with maximum activity of 14 U/ml at 30℃ after 48hrs of incubation period at 150 rpm. Isolate B3 produced the lipase at 37℃ consistently for five days with an average activity of 8.0 U/ml and this isolate may further be explored for industrial production of lipase as part of a dietary supplement for fat and oil metabolism. Although the isolate B1 lacked consistency in its lipase productivity, it may further be studied to retain the higher activity and produce the lipase consistently with the use of different substrates and optimized parameters.
Keywords: LipaseProducing Bacteria, Novel Strain, Biochemical Characterization, Soil Samples
Article Outline
1. Introduction
- We are in the situation challenging the usage of products and services derived from the biological sources. This is evident from the developments such as the detergents changed from the simple caustic soda to a mix of different enzymes of proteases &lipases[1,2] with stabilizers etc. It is also found that the varieties of dairy products like cheese, ice creams, butter milk, ghee etc. are produced with the use of lipase enzymes creating the opportunities and building the economies by ever increasing industry establishments all over the world[3,4]. The populations are also sensitizing not only to the variety of food products but also towards the quality of the products, thus bringing the variations in the pricing of the same product. In addition, variety of enzymes are also being used either directly or through the microbial populations for the treatment of many industrial waste rich in organic matter containing oily matter as well. Out of the many different enzymes used in bio-industries, lipase is one of the versatile groups of biocatalyst used to carry out various hydrolytic reactions in both aqueous and non-aqueous media including their interface. Lipases are isolated from various sources with wide range of substrate specificity and non-specificity to catalyze the reaction involving chemo, region and enantio-selective transformations. Lipases play a vital role in the manufacturing and services sector for the mankind. In a nutshell it is found that lipases are used in the processing of fats and oils, detergents and degreasing formulations, food processing, the synthesis of fine chemicals and pharmaceuticals, paper manufacture and production of cosmetics and pharmaceuticals, bakery, brewing, biofuels, leather processing and general cleaning[5-14]. Considering the ever increasing demand for the better lipases in the industry and the need to understand the mechanisms of lipase actions and the scope to research for novel sources of lipase[12] the present study has been carried out to isolate and characterize the novel lipase producing bacteria from soil samples. Soil is a rich source of many types of microbial strains which can provide (or revive) a particular group of microbial strains necessary for the degradation of different contaminants thrown in to the soil. Hence the soil samples can be used to isolate the novel strains that may be used as a part of the microbial pool for the production of lipase at research labs and industries.
2. Materials and Methods
2.1. Collection & Processing of Soil Samples
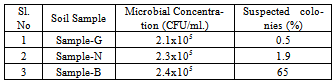
- Soil samples were collected from the out skirts of Guntur (Andhra Pradesh), Nagpur (Maharashtra) and Udaipur (Rajasthan), India; at a depth of 4-5 cm with the help of a sterile spatula in sterile plastic bags and brought to Microbiology laboratory, Department of Biotechnology, Sir PadampatSinghania University, Udaipur, Rajasthan, India; within 48 hrs. The soil sample from Guntur, Nagpur and Udaipur are named as Sample-G, Sample-N and Sample-B respectively. The samples were sealed in sterile containers and stored in the refrigerator at 4oC until use.
2.2. Screening of Lipase Producing Bacteria
- 1gm of soil from each sample was dissolved in 100 ml of 0.8% saline water separately and agitated for 30 min. in orbital shaker incubator at 30oC and 100rpm. Thus prepared soil suspension was used to make the dilutions of 10-1 to 10-6 for each of the soil samples by serial dilution method. These diluted soil samples of 1ml each were used as inoculum in tributyrin agar base medium for the screening of lipase producing bacteria by pour plate method. The prepared agar plates in duplicates were incubated at 30oC for 48 hrs. The colonies appeared were counted along with suspected lipase producing (with clear zone formation) bacteria. The total number of colonies in each soil sample was calculated [15] by the chosen consecutive third and fourth dilutions using the equation (1).
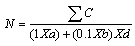 | (1) |
|
2.3. Selection of Best Isolates for Lipase Production
- The isolates capable of lipase production were further screened to isolate the best possible lipase producing bacteria based on agar well diffusion (Cup well method) and titrimetric (pH stat method) assays.
2.3.1. Shake Flask Culturing of Lipase Producing Isolates
- Liquid broth media was prepared with standard nutrient broth media containing 1% (v/v) tributyrin emulsified with homogenizer and autoclaving at 121oC & 15 psig for 15 min. All the lipase producing isolates were inoculated separately in to the prepared tributyrin broth media aseptically and incubated in shaker incubator at 150 rpm with different incubation temperatures ranging from 30oC to 50oC for five days. Crude lipase enzyme samples were extracted for every lipase producing isolate at an interval of 24hrs and assayed for lipase activity.
2.3.2. Lipase Crude Enzyme Preparation from Broth
- Microbial broth of 2ml were withdrawn separately from the broth culture of different lipase producing isolates at an interval of 24hrs for five days in a sterile vial aseptically and centrifuged at 10000g, 4oC for 10 min. in a cooling centrifuge. The supernatant was collected in a fresh vial and used it immediately as a lipase crude enzyme source for agar well diffusion & titrimetric assay of lipase.
2.3.3. Agar Well Diffusion Assay of Lipase Crude Enzyme
- Tributyrin agar base added with 1% (v/v) tributyrin was emulsified using homogenizer and autoclaved at 121oC, 15 psig for 15 min and cooled to 45oC under laminar air flow and poured in to the sterile glass petri plates aseptically. The agar media was let cool under laminar hood and closed the lid and kept inverted for 24hrs to obtain the sterile tributyrin agar plates. These sterile agar plates were punched aseptically with sterile cork borer to obtain two 4mm dia. wells in the two halves of the plates with approximate separation distance of 30mm. These tributyrin agar plates were loaded with freshly extracted crude lipase enzyme of 50 µl in each well separately for each extracted crude enzyme and incubated at 37oC in the incubator for 48 hrs. The developed clear zones around the wells were measured (mm) and the data was used for further analysis along with titrimetric assay data to screen the primary isolates for lipase producing bacteria.
2.3.4. Titrimetric Lipase Assay Method
- Lipase activity was measured by titrimetric method [16] using olive oil as substrate at pH 7.0. The reaction cocktail was prepared by 5% (w/v) olive oil emulsified in 5% (w/v) gum acacia in 100mM sodium phosphate buffer, pH 7.0. 1ml of each crude lipase enzyme was added to the reaction cocktail of 10ml separately at their respective culturing temperatures and incubated for 15 min. at 100 rpm in a shaker incubator. The reaction was quenched and fatty acids were extracted by adding 1ml of acetone: ethanol solution (1:1) and swirling the contents swiftly. Phenolphthalein indicator of 2-3 drops was added to each of the reaction mixture with respect to different crude lipase enzymes of isolates and the control. The contents of each reaction mixture were titrated with 0.05M NaOH solution to an end point of pink color at pH 10.0[17,18]. Lipase activity was calculated as micro moles of free fatty acids formed from olive oil per ml of crude lipase enzyme [11] as per equation (2).
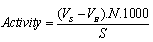 | (2) |
2.3.5. Screening of Isolates with Respect to Temperature
- The best lipase producing isolates screened from the soil samples were further cultured in tributyrin broth at 150 rpm in the shaker incubator at different incubation temperatures ranging from 30oC to 50oC and the broth samples were withdrawn separately at every 24hrs time interval to extract the crude lipase enzyme as described in the section 2.3.2 and estimated the lipase activity by titrimetric method for 5 days of culturing. Obtained data was used to screen the isolates for better lipase producing isolate(s) as shown in the Figures. 1-10.
2.4. Morphological and Biochemical Characteristics of the Best Lipase Producing Isolates
- Morphological studies of the best lipase producing isolates were studied under the light microscopy by gram staining method. The IMViC tests were performed for biochemical characterization.
3. Results & Discussion
- The soil Sample-B got higher number of microbial colonies (Table-1) with higher percentage of suspected colonies and isolates followed by Sample-N and Sample-G respectively, deducing direct proportionality relation between abundance of isolates and the number of viable microbial colonies in the soil samples. It is also interesting to note the same order of microbial content (105) with minor variation in the multiplication factor of the soil samples collected from three far away locations; which may be due to the similar climate conditions of the soil collected locations. Out of the suspected colonies isolated from the Samples-B, 12 isolates were found to be true lipase producing bacteria and from them five isolates were found to be higher lipase producing with nearly similar lipase activity profile. From Sample-G suspected colonies were isolated and from them 5 strains were found to be true lipase producing isolates; subsequently two isolates were screened to be the best isolates from Sample-G. From the suspected colonies of Sample-N, 5 strains were found to be true lipase producing isolates which have yielded one best isolate bringing preliminary best isolates to be eight from all the soil samples for further screening and analysis.
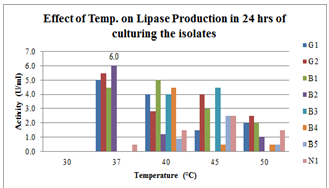 | Figure 1.Effect of Temperature on lipase production for 24hrs broth culturing of preliminary lipase producing isolates |
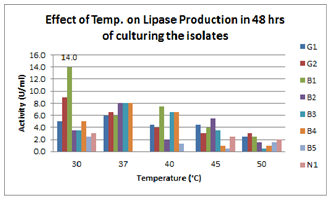 | Figure 2.Effect of Temperature on lipase production for 48hrs broth culturing of preliminary lipase producing isolates |
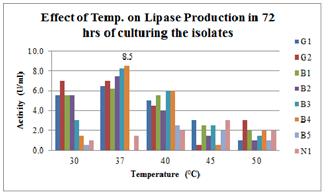 | Figure 3.Effect of Temperature on lipase production for 72hrs broth culturing of preliminary lipase producing isolates |
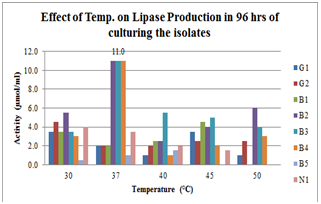 | Figure 4.Effect of Temperature on lipase production for 96hrs broth culturing of preliminary lipase producing isolates |
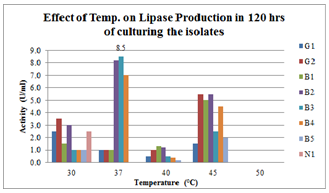 | Figure 5.Effect of Temperature on lipase production for 120hrs broth culturing of preliminary lipase producing isolates |
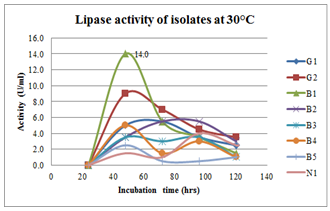 | Figure 6.Effect of incubation period on lipase production at 30oC inbroth culturing of preliminary lipase producing isolates |
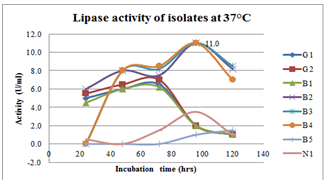 | Figure 7.Effect of incubation period on lipase production at 37oC inbroth culturing of preliminary lipase producing isolates |
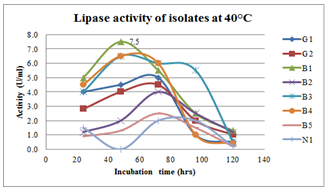 | Figure 8.Effect of incubation period on lipase production at 40oC inbroth culturing of preliminary lipase producing isolates |
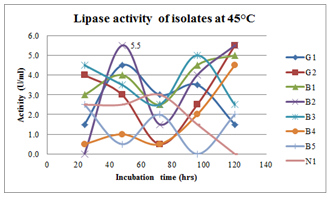 | Figure 9.Effect of incubation period on lipase production at 45oC inbroth culturing of preliminary lipase producing isolates |
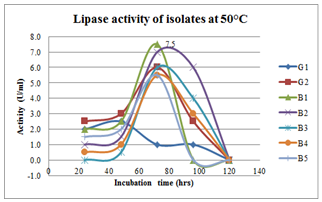 | Figure 10.Effect of incubation period on lipase production at 50oC inbroth culturing of preliminary lipase producing isolates |
5. Conclusions
- The isolate B1 was found to be the best possible isolate with highest lipase activity of 14 U/ml at 30oC with an incubation period of 48hrs among the screened eight lipase producing isolates. On an average the isolate B3 has produced the best lipase activity of 8.0 U/ml throughout the incubation period at 37oC which concludes the consistent and steady degradation rate of trybutyrine by lipase of isolate B3 and used the products for its growth and metabolic activities. Although it was found that 75% of the isolates produced an average of 8.0 U/ml of lipase activity for the culturing period between 48-72hrs at 37oC, the isolate B3 may be a better choice for the consistent production of stable lipase enzyme that may be of interest for its possible use as an ingredient in food supplement or digestion aid for human consumption[19]. The variation in the production of lipase by the isolates points towards their ability to tune the growth and metabolic activities with some time lag. It would be the interest of the pursuer to select the best isolate as per the production conditions and the intended use of the end product, lipase enzyme at its best activity. All the isolates were found to be the gram positive rod shaped bacteria. The selected best isolates can further be subjected to biochemical tests to identify and perform the optimization studies with respect to other physiological parameters for their possible exploration at industrial and lab scale production of lipase.
ACKNOWLEDGEMENTS
- The authors are grateful to Prof. P. C Deka, the Vice Chancellor, Sir PadampatSinghania University, Udaipur, Rajasthan, India, for his critical comments and insights at various stages of the research work. Dr.PallaviDwivedi, Assistant Professor, Department of Biotechnology, School of Engineering, Sir PadampatSinghania University, Udaipur, Rajasthan, India, is also gratefully acknowledged for the help and suggestions at various stages of the research work carried out.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-Text HTML
Full-Text HTML