-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Agriculture and Forestry
p-ISSN: 2165-882X e-ISSN: 2165-8846
2014; 4(5): 408-413
doi:10.5923/j.ijaf.20140405.10
Association of Xanthomonas sp. with Apple Trunk Canker Disease in an Apple Orchard from Iran
Nadjmieh Shaki1, Nader Hasanzadeh1, Mansoureh Keshavarsi2, Eisa Nazerian3
1Department of Plant Pathology, Faculty of Agriculture and Natural Resources, Science and Research Branch, Islamic Azad University, Tehran, Iran
2Seed and Plant Improvement Institute, Karaj, Iran
3Plant Protection Department, National Research Station of Ornamental Plants, Mahallat, Iran
Correspondence to: Nadjmieh Shaki, Department of Plant Pathology, Faculty of Agriculture and Natural Resources, Science and Research Branch, Islamic Azad University, Tehran, Iran.
| Email: |  |
Copyright © 2014 Scientific & Academic Publishing. All Rights Reserved.
During a survey of fruit orchards in northern part of Iran, some apple trees with trunk canker symptoms were observed. The infected trees lacked any visible symptoms on leaves and fruits. On NA and YDC media a Gram-negative, yellow-pigmented bacteria were consistently isolated. Based on pathogenicity, biochemical and molecular tests and fatty acid analysis, the bacterial agent was identified as Xanthomonas sp. The isolates were pathogenic on apple cv. prime gold detached leaves in laboratory conditions and in one-year old seedlings of cv. golden delicious in glasshouse condition. A typical canker symptoms were observed in apple trunks after seven months from artificial inoculation and the agent was re-isolated and Koch's hypothesis were fulfilled. DNA sequences were matched with Xanthomonas sp. sequence deposited in to NCBI GenBank. The bacterium could only survive and infect in high relative humidity and at temperature ranging 26-28°C. This is the first report of naturally occurring Xanthomonas sp. from apple canker disease symptoms.
Keywords: Trunk canker, Apple, Xanthomonas sp
Cite this paper: Nadjmieh Shaki, Nader Hasanzadeh, Mansoureh Keshavarsi, Eisa Nazerian, Association of Xanthomonas sp. with Apple Trunk Canker Disease in an Apple Orchard from Iran, International Journal of Agriculture and Forestry, Vol. 4 No. 5, 2014, pp. 408-413. doi: 10.5923/j.ijaf.20140405.10.
Article Outline
1. Introduction
- Apple (Malus domestica Borkh) is the third horticulture products in Iran, accounting 1662430 tons fruit product per year. Also, Iran is the 8th apple producer in the world (FAOSTAT, 2010). Many common bacterial diseases such as fire blight, blister spot, crown gall and hairy roothave been reported on apple worldwide (Vanneste, 2000 and Bradley et al., 2010) but natural association of apple trunk canker with Xanthomonas sp. has not been yet reported. Only a rare occurrence of an unusual Xanthomonas campestris strain with apple was observed on symptomatically apple tissue in vitro (Mass et al., 1985).Xanthomonas is a genus of Gammaproteobacteria that includes numerous phytopathogenic species, each characterized by a narrow host range. However, as a whole, the genus members are able to infect a broad range of plants, distributed over 124 monocotyledonous and 268 dicotyledonous plant species (Albuquerque et al., 2011). Some of the Xanthomonads could cause severe damage on economically important crops. Most of these pathogens have a well-defined and narrow plant host range (Maes, 1993). The genus also includes strains which may be associated with plant material but are not pathogenic (Brenner et al., 2005).In this study, bacterial isolates associated with natural trunk cankers on apples were isolated and investigated using some standard techniques.
2. Material and Methods
2.1. Isolation and Preliminary Characterization of Bacterial Isolates
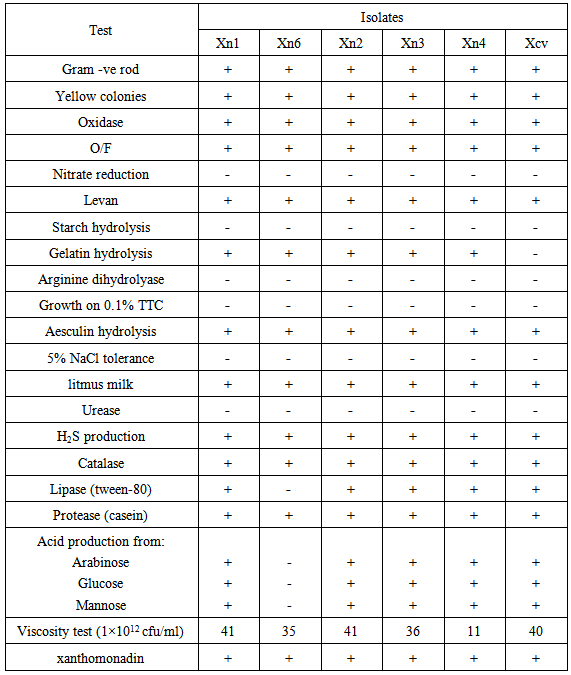
- During spring and summer 2010, an orchard located in the northern part of Iran was surveyed and about 30 canker samples on local apple cultivars were collected. Small pieces of trunk and branches were cut and immersed in 5 ml saline solution (0.80% NaCl) for 20 min and 30µl of suspension was cultured on nutrient agar (NA) and incubated at 26°C for 3-5 days. Yellow pigment colonies were purified and subjected to key physiological and biochemical tests including gram reaction (Suslow et al., 1982), fluorescent pigment production (King et al., 1954), growth at 25°C, 27°C and 37°C, oxidase (Kovacs, 1956), catalase (Bradbury, 1986), Oxidative ⁄ Fermentative (O/F) (Hugh and Leifson, 1953), levan production from sucrose (Lelliott and Stead, 1987), gelatin and starch hydrolyses (Fahy and Persley, 1983), nitrate reduction, production of arginine dihydrolase, ability to produce acid from carbohydrates (arabinose, glucose and mannose), growth in presence of 5% NaCl, proteolytic, lipolytic and amylolytic capability, aesculin hydrolysis (Schaad et al., 2001), Xanthomonadin production (Starr et al., 1981) and viscosity test (Klement, 1990) and all key biochemical tests were done in dual cultures (Gracelin et al., 2012). A dendrogram based on these results was constructed using the NTSYS-pc.
2.2. Hypersensitive Reaction (HR)
- The bacterial suspensions were prepared from pure cultures on NA medium and their optical density was adjusted spectrophotometerically to 107 cfu /ml. Using a hypodermic syringe, each bacterial suspension was injected into fully expanded tobacco cv. White Burley and geranium cv. Pelargonium Xhontorum Bailey leaves and visible necrosis symptoms in the inoculated areas were recorded from 24-48 hours post inoculation (Fahy & Persley, 1983).
2.3. PCR Amplification and Sequencing of 16S rDNA
- Bacterial DNAs of five strains of apple (Xn1, Xn2, Xn3, Xn4 and Xn6) and Xanthomonas campestris pv. vesicatoria (Xcv) were extracted by alkaline lysis method. Colonies on agar plates were suspended in 10 µl of lysis buffer (100 µl of 0.05 M NaOH was added to 10 μl of cell suspension and incubated at 95°C for 15 min). The bacterial suspensions were centrifuged for 2 min at 14,000 rpm. 1 µl from each culture supernatant was used for each PCR reaction. DNA segments were then amplified with reverse primer of 8/27 (5′-AGAGTTTGATCITGGCTCAG-3′) and forward primer of 461/477 (5′-AAGGATCGGGTATTAAC-3′) (Maes, 1993; Palacio-Bielsa et al., 2009) and the products were sequenced and compared with sequences available in the NCBI GenBank databases (http://www.ncbi.nlm.nih.gov).
2.4. Pathogenicity Tests
2.4.1. Detached Shoot Test
- Young shoots of apple cv. prime gold were surface sterilized and inoculated with all 17 bacterial suspensions of Xanthomonas sp. and Xcv (as well as an authentic culture of X. vesicatoria). Each treatment was replicated three times. The inoculated shoots were kept standing in sterile water covered with polythene bags for 48 hours. All treated shoots were incubated in a growth chamber at 26°C for 7-10 days. The development of any visible symptoms were compared and recorded (Lelliott and Stead, 1987).
2.4.2. Greenhouse Test
- The five strains of Xn1, Xn2, Xn3, Xn4 and Xn6 with potential pathogenic traits were chosen for this confirmatory test. The trunks of two-years-old seedlings of apple cv. Golden delicious in four replicates were inoculated with above-mentioned bacterial suspensions. The wounds were covered with moist cotton and a strip of parafilm for 48 hours. The treated seedlings were maintained in a greenhouse at 25-27°C and 95% relative humidity. The reaction of the seedlings was observed weekly for about 8 months (Santi et al., 2004).
2.5. Fatty Acid Analysis
- Whole cell fatty acids were analyzed using GC-Mass (Massomo et al., 2003, Sasser, 2001 and Vauterin et al., 1996). Bacteria were streaked on tryptic soy broth agar and grown at 26°C for 48 h. A loopful of bacteria was added to a 13×100 mm screw-cap tube. One ml of reagent 1 (45g sodium hydroxide, 150 ml methanol in 150 ml distilled water) was added to each tube containing bacterial cells. The tubes were then securely sealed with teflon lined caps, vortexed briefly and heated in a boiling water bath for 5 minutes. This was repeated once more and heat treatment was completed in 30 minutes. The samples were cooled to room temperature and methylated with 2ml of reagent 2 (325ml 6.0M hydrochloric acid and 275 ml methyl alcohol, pH<1.5). The tubes were capped and heated for 10 ± 1minutes at 80°C and cooled to room temperature. Fatty acid methyl esters were extracted by adding 1.25ml of reagent 3 (200ml hexane and 200ml methyl tert-butyl ether) to the cooled tubes followed by recapping and gentle tumbling on a clinical rotator for 10 minutes. The aqueous (lower) phase was pipetted out and discarded. Samples were washed by mixing 3ml of the reagent 4 (10.8g sodium hydroxide dissolved in 900 ml distilled water). The organic top phase was taken into tubes, recapped and tumbled for 5 minutes. About 2/3 of the organic phase was pipette into a GC vial and capped for analysis. GC analyses were carried out using a Hewlett Packard (HP) series 6840 gas chromatograph, equipped with 30-m-long capillary column and a mass-selective detector (model HP 5973).
3. Result and Discussion
3.1. Isolation and Preliminary Characterization
- In total 17 bacterial strains with small, yellow and mucoid colonies were isolated. Five strains (Xn1, Xn2, Xn3, Xn4 and Xn6) with typical morphological characteristics were further characterized by phenotypic and molecular methods. Results of the key biochemical tests were summarized in Table 1 and converted into dendrogram using NTSYS (Figure 1). All five isolates were shown a peak at 420-450 nm characteristic of xanthomonadin pigment.
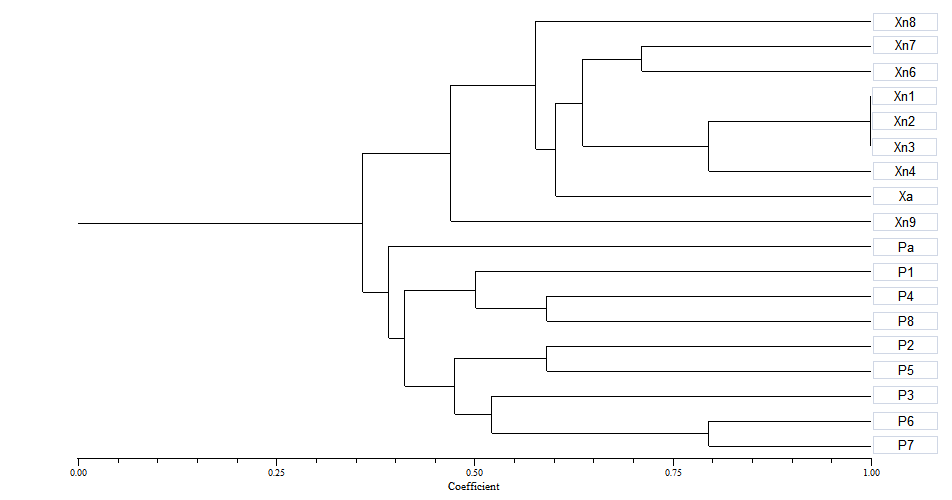 | Figure 1. UPGMA dendrogram showing the phenotypic similarities of 17 apple strains and isolate of X. vesicatoria (Xcv). Alomost all apple strains (Xn1-4 and Xn5-8) plus Xcv were located in cluster 1 |
|
3.2. Determination of the 16S rDNA gene Sequences
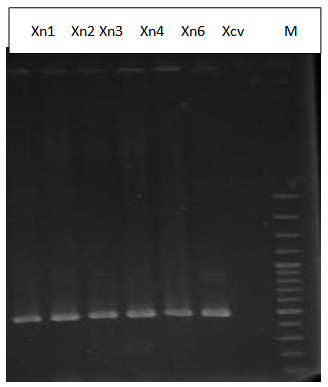
- All 5 Xanthomonas strains including Xcv produced a single sharp 480-bp fragment (Figure 2). This result was coincided with initial PCR results (Maes, 1993) and also in accordance with known phenotypic characteristics of Xanthomonads.The phylogenetic affiliation of the five strains were also determined. Sequence analysis of strains were indicating these were belonged to the genus Xanthomonas. 16S rDNA gene sequence of the isolate Xn1 has been deposited to GenBank with accession number of JF920121.
3.3. The Hypersensitive Reaction
- Typical necrosis was developed in inoculated sites of geranium but not on tobacco leaves 48 h after inoculation (Figure 3).
 | Figure 3. Positive hypersensitive reaction symptoms on geranium (left) but not on tobacco leaves (right) |
3.4. Pathogenicity Tests
- Wilting and dieback symptoms were observed within 7 days on treated young detached apple shoots. In greenhouse assay the treated seedlings undergone gradual bark discoloration and wilted in eight-month period. The control seedlings were remained healthy (Figure 4).
 | Figure 4. Wilting of detached apple shoot (left) and bark discoloration and wilting of entire seedling (right) in greenhouse condition |
3.5. Fatty Acid Analysis
- No satisfactory result was obtained from fatty acid analysis. However, few fatty acids characteristic of the genus Xanthomonas were detected in two tested strains. Composition of fatty acids for representative apple strain (Xn1) was 10:0, 15:0, 16:1 9cis, 16:1 anteiso, 18:0 and 18:1 9cis. This was 14:1, 15: 0, 14: 0, 16:1 9cis, 16:1 anteiso and 24:0 for authentic culture of Xcv.
4. Conclusions
- Based on certain criteria such as biochemical tests, xanthomonadin and fatty acid analysis and a molecular method, the five representative strains of a plant-associated xanthomonad were tentatively identified as Xanthomonas sp. Blast analysis with the sequences of strain Xn1 confirmed that this novel bacterium was belonged to the Xanthomonas group of the γ-Proteobacteria with highest similarity 98% with the genus Xanthomonas. No significant differences could be found among the strains of the cluster 1 accommodating all five pathogenic strains of Xanthomonas sp.To the best of our knowledge no pathogenic Xanthomonas sp. has been yet reported on apple trees under natural field condition. The only record in this area is the association of unusual strain of X. campestris with damaged apple explants in tissue culture materials (Mass et al., 1985). In the latter, they failed to find any relationship between Xanthomonas campestris and apple host plants and all their attempts to fulfill the pathogenicity tests on plants such as tomato, tobacco, cassava, peach and grape were disappointed. In fact only a limited number of plant pathogenic bacteria were found causing diseases on apple trees (Bradley et al., 2010). Results of pathogenicity tests and variation in symptom developments were indicated that this novel bacterium is capable to survive, multiply and infect apple trunk in natural condition for a prolonged periods of time. But neither of strains could survive on leaf/bud surfaces. The techniques used for characterization of this bacterium did not precisely identify the isolates at species level. To achieve this, more confirmatory screening tests particularly more molecular methods, fatty acid profiles and cultivar reactions to single/mixed cultures of the isolates were required.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML