-
Paper Information
- Next Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Agriculture and Forestry
p-ISSN: 2165-882X e-ISSN: 2165-8846
2014; 4(1): 41-45
doi:10.5923/j.ijaf.20140401.06
Diversity among Bacterial Isolates of Pectobacterium and Dickeya Causing Soft Rot of Ornamental Plants in Western Mazandaran, Iran
SaharSayad1, Nader Hassanzadeh1, Eisa Nazerian2, Abolghasem Ghasemi3
1Islamic Azad University, Science and Research Branch, Tehran, Iran
2Ornamental Plants Research Institute, Mahalat, Iran
3Iranian Research Institute of Plant Protection,Tehran, Iran
Correspondence to: SaharSayad, Islamic Azad University, Science and Research Branch, Tehran, Iran.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
Bacteria cause soft rot on a wide range of crops. The isolates involved in soft rot of many ornamental plants have been researched extensively due to their economic importance. Based on the phenotypic features and results of the molecular methods, it was evident that Pectobacteriumspp and Dickeya spp are the main bacterial soft rot causal agent associated with ornamental plants in Western Mazandaran, Iran. In the present study 26 isolates were investigated for detection and determination of diversity among isolates of Pectobacterium and Dickeya. The variation within the isolates was studied in terms of physiological characteristics, pathogenicity and molecular methods in this paper P. betavascularum is reported on Aglaonema for the first time.
Keywords: Detection, Identification, Pectobacterium, Dickeya, PCR
Cite this paper: SaharSayad, Nader Hassanzadeh, Eisa Nazerian, Abolghasem Ghasemi, Diversity among Bacterial Isolates of Pectobacterium and Dickeya Causing Soft Rot of Ornamental Plants in Western Mazandaran, Iran, International Journal of Agriculture and Forestry, Vol. 4 No. 1, 2014, pp. 41-45. doi: 10.5923/j.ijaf.20140401.06.
1. Introduction
- Most Pectobacterium which cause soft rot, lack specificity of attacking the hosts, while certain Pectobacterium species has a narrow range of crops. The five soft rot pathogens of Pectobacterium: P. atrosepticum, P. betavascularum, P. wasabiae, P. carotovorum, and P. odoriferum have been thoroughly examined taxonomically[25,13,12]. Pectobacterium species are distinguished by their ability to produce large quantities of pectic enzymes (mainly pecticlyases) that enable them to cause soft rot in macerated parenchymatous tissue of a wide range of plant species. One of the famous bacterial diseases in potato is basal stem rot, also known as blackleg, which is associated with decay of the parent seed tuber. In many other crops, wilting and yellow leaves in early stages of attack may take place. In advanced stages chlorotic, necrotic and water soaked areas will occur. However, the symptoms of soft rot caused by Pectobacterium spp. in susceptible hosts under field conditions are indistinguishable. Pectobacterium produce extracellular enzymes such as proteases, polygalacturanase, pectatelyase which degrade the plant cell wall and membrane compounds lead to plant death[3,27]. Some chemicals have been proved for soft rot disease control but these have limitation in soft rot control[20,18]. Agricultural practices such as rotation, sanitation, removal of plant debris, appropriate plant nutrition, insect control, warehouse temperature control, use of clean irrigation water and planting of resistant cultivars and production of planting material free from pathogen by tissue culture technique, soft chemicals, natural chemicals, disinfectants, calcium application, growth regulators, chemical elicitors to induce natural host defenses, biological control, integrated control, hypobaric pressure, physical means such as ultraviolet illumination, radiation, hot water or heat shock treatments, modified atmosphere storage and packaging genetic modification of plants[4] are effective means for Pectobacterium soft rot control. To-date several methods had been developed for detection of soft rot Pectobacterium. Selective growth media[26,24], immunological[30] and PCR based techniques[32,8] are more reliable methods used in many laboratories for Pectobacterium characterization. However, there are some specific probes[6] for Pectobacterium characterization, but in routine tests they are limited by low signals when labeled with non-radioactive probes. A set of primer designed to amplification of 434-bp fragment of a pectatelyase encoding gene (pel gene) in order to detect and identify all Pectobacterium species (atrosepticum, carotovorum, odoriferum, and wasabiae), except betavasculorum[6]. Another pair of primers (EXPCCR and EXPCCF) was also designed to amplify a 550bp fragment in P. carotovorum and P. wasabiae strains. Digestion of obtained PCR product with Rsaɪ restriction enzyme represented 5 different RFLP patterns in agarose gel[15]. a banding pattern of 620-bp was produced from pure cultures of P. atrosepticum using Eca1f and Eca2r primers[7]. Powerful analytical tools (e.g. specific genes and DNA sequencing, DNA-DNA hybridization, isozyme analysis, randomly amplified polymorphic DNA, restriction fragment length polymorphism (RFLP) and rep-PCR are used for identification and differentiation of isolates even at strain level, which are important for ecological and epidemiological surveillance purposes. These procedures require only a small proportion of samples for examination. The discriminatory power of these markers is very high and closely related strains can be differentiated. The occurrence of soft rot bacterium of ornamental plants have been previously reported in northern Iran[16,2]. The aims of this study were detection and determination of genetic diversity of some soft rot isolates collected from ornamental plants using biochemical and PCR-based techniques.
2. Materials and Methods
- Leaves and stems of ornamental plants (table 1) infected by soft rot bacterium with water soaked lesions, wilting, and soft rot symptoms were collected randomly from 50 commercial ornamental greenhouses in northern Iran during 2009-2010. In total, 60 samples were collected and selected for characterization assays. Infected tissues were surface sterilized using 70% ethyl alcohol. Small pieces of infected tissues were soaked in saline solution (0.85% NaCl) for 20 min to disperse the bacterial cells into the solution. Standard protocol using eosin methylene blue (EMB), nutrient agar (NA) and King's B (KB) media were used for isolation of bacteria from the plant samples[30,7]. Twenty six pure cultures were collected and stored at -80°C. The authentic cultures for Pectobacterium atrosepticum 1043 SCRI, Dickeya dadanticola 3937 SCRI (SCRI Scottish Crop Research), Dickeya dianthicola IPO 980, Dickeya sp. IPO 2222, Pectobacterium carotovorum subsp. carotovorum IPO 1949, Dickeya chrysanthemi DSM 4610 (German Resource Center for Biological Material) were used as standards. Isolates which were positive to pectolytic activity selected for the conventional biochemical and physiological tests including facultative anaerobic activity, gram reaction, erythromycin sensitivity, iodole production, oxidase, phosphatase, malonate utilization, reducing substance from sucrose, methyl red, nitrate reduction, growth on 5% sodium chloride (NaCl), lecithinase, urease and arginine dihydrolayse, casein, starch, Tween hydrolyses, H2S production and assimilation of carbon sources using basal medium of Ayers[30]. Plants inoculation was done by injection of 108 CFU/ml bacterial suspensions from 24h slant culture into stem of young ornamental plants and incubated at 28°C with 85-90% relative humidity[10,1]. Controls were treated only with sterile water. Four primer pairs ADE1/ ADE2, Y1/Y2, EXPCCR/EXPCCF and ECA1f/ECA2r were used to detect Dickeya and Pectobacterium isolates in PCR assay[6,8,22,15]. PCR was performed in 25µl of reaction mixture. Amplified DNA fragments were run on 1.5% agarose gel at 90V for 1h and visualized under UV-Light following ethidium bromide staining. In rep-PCR analysis total bacterial genomic DNAs were extracted using Alkalin Lysis Method[28]. Colonies on agar plates were suspended in 10 µl of lysis buffer (100 µl of 0.05 M NaOH was added to 10 μl of cell suspension and incubated at 95°C for 15 min). The bacterial suspensions were then centrifuged for 2 min at 12,000 rpm. Genomic DNA (50ng/µl) was used as the template for all rep-PCR reactions[14]. Two primers of BOX and ERIC were used as described by Rademaker et al.[28]. The amplification cycles were consisted of 35 cycles for denaturation (94°C/1min), annealing (48°C/1min), extension (72°C/2min) and final extension (72°C/10min). Amplified DNAs were analyzed by electrophoresis in a 1.5% agarose gel in TBE 0.5X buffer. A 3kbp ladder was used as a size marker. Banding patterns were stained with ethidium bromide and visualized under UV light. To determine the relationships among strains, cluster analysis was made by UPGMA algorithm with the use of NTSYS software pc2.0[5]. Banding patterns were recorded using UviDoc® (version 99.02) software and bands were scored as '1' for presentand '0' for absence. The dendrograms were constructed from a similarity matrix using Dice’s coefficient [29].
3. Results and Discussion
- In total, twenty six isolates with cream-white colonies were purified from soft rotted plant tissues. Based on certain characteristics such as positive for indole production and phosphatase activity, reducing substances from sucrose, sensitive to erythromycin, positive to produce soft rot on potato and hyper sensitivity reaction on tobacco, able to growth in 5% NaCl, reduction of catalase and nitrate and production of H2S from cystein, the isolates were identified as either Pectobacterium spp. or Dickeya spp. The Pectobacterium strains were further divided into three groups based on bacteriological characteristics. Seven strains belonging to group I, was negative for casein, VP, and indole production, as well as phosphatase activity, citrate utilization and lecitinase test. This group was clustered in single group of Pectobacterium carotovorum. One strain belonging to group II (Pectobacterium atrosepticum) was negative for casein, and VP production and growth at 36°C and positive to α-methyl glocoside, citrate and MR tests. The group III with single strain of Pectobacterium betavascularum gave negative response to casein, citrate and MR tests and was positive for VP. Three strains (11, 26, and 28) also were identified as Dickeya chrysanthemi and placed in a separate group. The Microlog system was also employed to identify two isolates (13, 15) as Pectobacterium betavascularum and isolate 11 as Dickeya chrysanthemi with similarity index of 67%, 49% and 63%, respectively. Inoculation of representative bacterial suspension into ornamental plants parts produced typical bacterial soft rot symptoms. The rotted tissues with mushy appearance were observed 2-5 days after inoculation. No symptoms were developed on control plants. A 434bp PCR-amplified fragments corresponding to pectatelyase-encoding gene was obtained in ten strains of Pectobacterium spp. by using Y1/Y2 primers. After PCR amplification with ECA1f/ECA2r primers, one isolates (19) produced a 690bp banding pattern designated to be P. atrosepticum. Likewise, the primer set (EXPCCF/ EXPCCR) amplified a single fragment of the expected size (550bp) for seven isolates of Pectobacterium carotovorum (36, 9, 8, 7, 5, 3, 1) an expected band of 420bp was also obtained from three Dickeya strains using ADE1/ADE2 primers. ERIC-PCR and BOX-PCR analysis were produced different fingerprinting profiles based on their origin and the primers used. The use of the PCR based methods for characterization of P. carotovorum subsp. carotovorum was strongly depicted with primers analogous to ubiquitous repetitive DNA sequences[9] to generate specific DNA fingerprints of Pectobacterium spp. ERIC-PCR and BOX-PCR analysis, showed that, bacteria isolated from different hosts gave different fingerprinting profiles, based on the primer used, the patterns of the reference isolates were different from those of isolates obtained from different plants, but identical in both methods. An UPGMA dendrogram constructed using the patterns generated with the two primers and the Dice index values confirmed that the isolates from ornamental plants were identical. The DNA fragment sizes varied between about 190 to 3800 bp with the use of the BOX primer. Both methods, showed observable diversity among isolates from every species of ornamental plants. Multiple and different DNA fragments yielded by amplification of chromosomal DNA with BOX and ERIC primers, revealed that there was divergence among isolates. Generally, the groups generated by ERIC-PCR and BOX-PCR are related to the plant species and geographical origins. Because of this, most of our isolates from specific areas were placed in the main groups. The influence of geographical origins on bacterial genetics was reviewed before[31,19]. Undoubtedly, rep-PCR has high reliability in epidemiological studies of bacterial diseases and the information derived from it, is applicable for detection and genetically characterization of bacterial pathogens. Based on BOX-PCR results, two main clusters were formed at 65% similarity. The first cluster including Pectobacterium spp. sub-divided into three subgroups with 68% similarity. The second cluster identified as Dickeya spp. divided into two subgroups with 71% similarity (figure 1). In ERIC-PCR analysis the similarity values between two main clusters was 52%, between subgroups of Pectobacterium spp. 59% and among Dickeya spp. subgroups was 70% (figure 2). From twenty six strains that were presumptively identified as Pectobacterium spp., ten were confirmed as Pectobacterium species and three Dickeya chrysanthemi using biolog and PCR methods. BiologMicrolog® system also identified isolates 13 and 15 as Pectobacterium betavascularum and 11 as Dickeya chrysanthemi. Molecular fingerprints based on BOX-PCR and ERIC-PCR, showed large genetic diversity within strains of Pectobacterium spp. The reason for this variability is unknown to many authors but might be related to some environmental and host conditions such as different levels of relative humidity and temperature and nutrition resources in greenhouses[4]. We here applied PCR-based technique with specific primers with this aim to increase detection sensitivity, simplicity and rapidity compared with phenotypic and biochemical assay. This led to accurate identification of Pectobacterium carotovorum, Pectobacterium atrosepticum and Dickeya chrysanthemi. However, thirteen strains with identical characteristics of the genus Pectobacterium failed to be amplified with specific bands. In this study, close correlations between biochemical characteristic and BOX-PCR based genetic finger prints among some of the strains were observed. The strains of similar genetic groups were exhibited similar phenotypic features and thus were placed in the same genetic/phenotypic group. Some similarities and differences were obtained from BOX-PCR and ERIC-PCR analyses. BOX primer grouped the strains more properly than ERIC primer and clustered P. carotovorum strains into 2 groups. Based our knowledge this is first report of bacterial soft rot on Aglaonema sp. (Chinese evergreen) caused by Pectobacterium betavascularum in Iran.
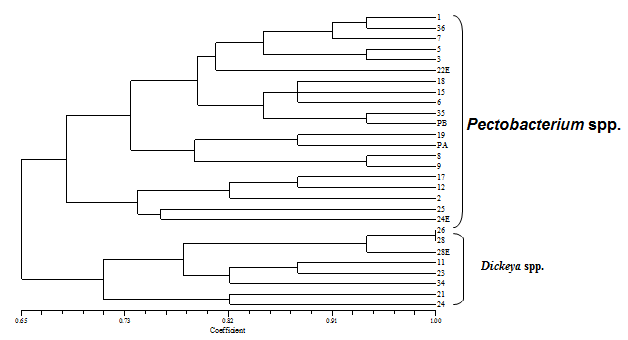 | Figure 1. Dendrogram based on BOX-PCR of isolates from ornamental plants |
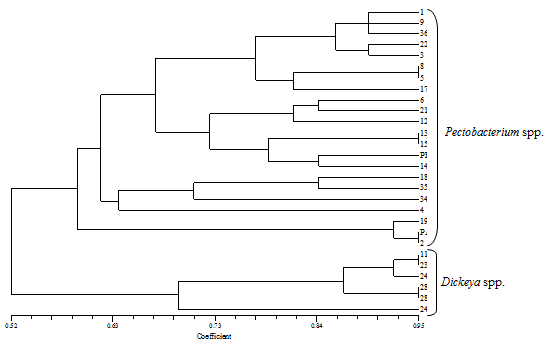 | Figure 2. Dendrogrambased on ERIC-PCR fingerprint profiles |
References
| [1] | Azad, H.R., Holmes, G.J. & Cooksey, D.A. 2000. a new leaf blotch disease of Sudangrass caused by Pantoe aananas and Pantoea stewartii. Plant Disease, 84,973- 97. |
| [2] | Baghaee-Rvari, S., Rahimian, H., Shams-Bakhsh, M., Lopez-Solanilla, E., Antunes-Lamas, M., and Rodriguez- Palenzuela, P., 2010. Characrerization of Pectobacterium speciese from Iran using Biochemical and Molecular Methods. European Journal Plant Pathology., 129, 413-425. |
| [3] | Barras, F., Gijsegam, V. F., Chatterjee, A. K., 1994. Extracellular enzymes and pathogenesis of soft rot Erwinia. Annual Review Phytopathology, 32, 201-234 |
| [4] | Costa, V. M., MCGrann, K. M., Hughes, D. W., Wright, G. D. 2006. Sampling the Antibiotic Resistance. Science, 311, 374-377. |
| [5] | Czajkowski, R., Grabe, G. J., and Vander Wolf, J. M., 2009. Distribution of Dickeya spp. and Pectobacterium carotovorum subsp. carotovorum in naturally infected seed potatoes. European Journal Plant Pathology, 125, 203-275. |
| [6] | Darrasse, A., Priou, S., Kotoujansky, A., and Bertheau, Y., 1994. PCR and reaction fragment length polymorphism of a pel gene as a tool to identify Erwinia carotovora in relation to potato diseases. Applied and Environmental Microbiology, 60, 1437-1443. |
| [7] | De Boer, S. H., and Kelman, A., 2001. Erwinia soft rot group. In: Laboratory Guide for identification of plant pathogenic Bacteria. 3rd ed., (N. W. Schaad, J. B. Jones, W. Chun, ed). American Phytopathological Society, St. Paul, MN, USA, 378pp. |
| [8] | De Boer, S. H., and Ward, L. J., 1995. PCR detection of Erwinia carotovora subsp. atroseptica associated with potato tissue. Phytopathology, 85, 854-858. |
| [9] | De Bruijn, F. J., 1992. Use of repetitive (repetitive extragenic palindromic and Enterobacterial repetitive intergenic consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl. Environ. Microbiol., 58, 2180-2187. |
| [10] | Dickey, R. S., Claflin, L. E., and Zumoff, C. H. (1987). Erwinia chrysanthemi: serological comparisons of strains from Zea mays and other hosts. Phytopathology, 77, 426–430. |
| [11] | Dickey, R. S., and Kelman, 1988. Erwinia "carotovora" or soft rot group. In Laboratory Guide for Identification of Plant Pathogenic Bacteria. (N.W. Schaad, ed). Amirican Phytopathological Society, ST. Paul, MN, USA, 44-59. |
| [12] | Duarte, V., De Boer, S. H., Ward, L. J., and de Oliveira, A. M. R., 2004. Characterization of atypical Erwinia carotovora strains causing black leg of potato in Brazil. Journal of Applied Microbiology, 96, 535-545. |
| [13] | Gardan, L., Gouy, C., Christen, R., and Samson, R., 2003. Elevation of three subsp. Of Pectobacterium carotovorum to species level: Pectobacterium atrosepticum sp. Nov., Pectobacterium betavascularum sp. Nov. and Pectobacterium wasabiae sp. Nov. International Journal of Systematic and Evolutionary Microbiology, 53, 381-391. |
| [14] | Kaneshiro, W.S. 2008. Characterization of Erwinia chrysanthemi from Bacterial Heart Rot of Pineapple Outbreak in Havaii. Plant Disease. 92(10), 00-00. |
| [15] | Kang, H. W., Kwon, S. W., and Go, S. J., 2003. PCR-based specific and sensitive detection of Pectobacterium carotovorum subsp. carotovorum by primers generated from a URP-PCR fingerprinting-derived polymorphic band. Plant Pathology, 52, 127-133. |
| [16] | Khalemlou, E., 2007. Determination of the taxonomic status of common and atypical isolates of Pectobacterium from several hosts in the Mazandaran, Iran, Dissertation, University of Tarbiat Modares, Tehran, Iran. |
| [17] | Lelliott, R. A., and Stead, D. E., 1987. Methods in Plant Pathology. Volume 2. Methods for Diagnosis of Bacterial Disease of Plants. British Society for Plant Pathology, Blackwell Scientific Publications, Oxford, UK. 216 pages. |
| [18] | Letal, J. R., 1977. Efficacies of disinfectants against potato ring rot and blackleg bacteria. American Potato Journal, 54, 405-409. |
| [19] | Louws, F. J., and Cuppels, D. A., 2001. Molecular techniques. Pages 321-337 in: Laboratory Guide for Identification of Plant Pathogenic Bacteria, 3rd ed. N. W. Schaad, J. B. Jones, and W. Chun, eds. American Phytopathological Society Press, St. Paul, MN. |
| [20] | Lund, B. M. and Lyon, G. D. 1975. Detection of inhibitators of Erwinia carotovora and Erwinia herbicola on thin layer chromatograms. Journal of Chromatography, 110:193-196. |
| [21] | Mahmoudi, E., Soleimani, M. J and Taghavi, M. 2007. Detection of bacterial soft rot Crown imperial caused by Pectobacterium carotovorum subsp. carotovorum using specific primers. Phytopathology Mediteranian, 46, 168-176. |
| [22] | Nassar, A., Darrasse, A., Lemattre, M., Kotoujansky, A., Dervin, C., Vedel, R., and Bertheau, Y., 1996. Characterization of Erwinia chrysanthemi by pectolyticisozyme polymorphism and restriction fragment length polymorohism analysis of PCR- amplified fragments of pel genes. Applied Enviromental Microbiology, 62, 2228-2235. |
| [23] | Norman, D. J., Yuen, J. M. F., Resandiz, R., and Boswell, J., 2003. Characterization of Erwinia population from nursery retention in Florida. Plant Disease, 87, 193-196. |
| [24] | Perombelon, M. C. M., and Burnett, E. M., 1991. Two modified crystal violet pectate (CVP) media for the detection, isolation and enumeration of soft rot erwinias. Potato Research, 34, 79-85. |
| [25] | Perombelon, M. C. M., and Kelman, A., 1980. Ecology of the soft rot Erwinias. Annual Review of Phytopathology. 18, 361-387. |
| [26] | Perombelon, M.C.M. & Hyman, L.J. 1986.a rapid method to identify and quantify soft rot erwinias directly from plant material based on their temperature tolerance and sensitivity to erythromycin. Journal of Applied Bacteriology, 60, 61-66. |
| [27] | Perombelon, M.C.M., Salmond, G.P.C. 1995. Bacterial soft rots in singh US, singh R.P, Kohmoto, K(Eds). Pathogenesis and host specificity in plant disease. Pergamon Press Ltd, Oxford, UK.1:1-20. |
| [28] | Rademaker, J.L.W., De Bruijn, F.J. 1997. Characterization and classification of microbes by rep-PCR genomic fingerprinting and computer-assisted pattern analysis. Pages 151-171 in: DNA Markers:Protocols, Applications and Overviews. G Caetoan-Anolles and P.M. Gresshoff. Eds.John wiley & Sons, Inc., New York. |
| [29] | Rolph, F. J., 2000. NTSYSPC Numerical taxonomy and multi variate analysis system. Version 2.7 Exter Software Applied Biostatistics Inc., NEW YORK, USA. |
| [30] | Schaad, N.W., Jones, J.B., Chun, W. 2001. Laboratory guide for identification of plant pathogenic bacteria (3rded). The American phytopathological Society, St Paul, Minn: APS., 373pp. |
| [31] | Scortichini, M., Rossi, M. P., 1991. In Vitro Susceptibility of Erwinia amylovora (Burrill) Winslow et al. to geraniol and citronellol. Journal of Applied Bacteriology, 71, 113-118. |
| [32] | Smid, E. J., Jansen, A. H. J., and Gorris, L. G. M., 1995. Detection of Erwinia carotovora subsp. atroseptica and Erwinia chrysanthemi in potato tubers using polymerase chain reaction. Plant Pathol., 44, 1058-1069. |
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML