-
Paper Information
- Next Paper
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Agriculture and Forestry
p-ISSN: 2165-882X e-ISSN: 2165-8846
2013; 3(4): 152-158
doi:10.5923/j.ijaf.20130304.05
Root-Associated Microorganisms Prevent Caffeine Accumulation in Shoots of Salvia officinalis L
Margot Schulz1, Mona Knop1, Carmen Muellenborn2, Ulrike Steiner3
1IMBIO (Institut für Molekulare Physiologie und Biotechnologie der Pflanzen), Universität Bonn, Bonn, 53115, Germany
2IBG-2, Plant Sciences, Forschungszentrum Jülich GmbH, Jülich, Germany
3INRES (Institut für Nutzpflanzenwissenschaften und Ressourcenschutz), Universität Bonn, Bonn, 53115, Germany
Correspondence to: Margot Schulz, IMBIO (Institut für Molekulare Physiologie und Biotechnologie der Pflanzen), Universität Bonn, Bonn, 53115, Germany.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
Salvia officinalis, previously used as an intercrop in coffee plantations, absorbed caffeine from incubation media. Caffeine was mainly found in the roots, together with traces of theobromine, while solely a small amount of caffeine was found in the shoots. In sterile plants, the caffeine accumulation was similar in both the roots and the shoots. The addition of antibiotics to the incubation medium of non-sterile sage revealed an involvement of plant-associated microorganisms in the caffeine degradation. Three most active microorganisms were isolated from non-sterile sage roots and were identified by molecular and microscopic studies as Trichoderma hamatum (fungus), Pseudomonas putida (bacterium) and Rhodotorula glutinis (fungus). Whereas P. putida and R. glutinis were associated with the rhizosphere, T. hamatum existed as an endophyte inside the roots, as ascertained by colonization and re-isolation studies. The study demonstrates that the degradation of caffeine is initiated by the ability of the microorganisms to perform demethylations, whereas the xanthine degradation may be attributed to either the plant or the microorganisms. Plants, which do not contain caffeine as a secondary product, have not yet been investigated for degradation of the compound. Trichoderma hamatum was not known as a caffeine degrading species and as an endophyte of Salvia officinalis. We propose a novel eco-biochemical interaction betweenendophytic T. hamatum and sage plants in caffeine degradation. If aromatic plants are used as intercrops in coffee plantations, the occurrence of caffeine degradation in those plants is of great importance.
Keywords: Allelopathy, Endophyte, Caffeine Degradation, Plant-fungus Interaction, Pseudomonas Putida, Rhodotorula Glutinis, Salvia Officinalis L., Trichoderma Hamatum
Cite this paper: Margot Schulz, Mona Knop, Carmen Muellenborn, Ulrike Steiner, Root-Associated Microorganisms Prevent Caffeine Accumulation in Shoots of Salvia officinalis L, International Journal of Agriculture and Forestry, Vol. 3 No. 4, 2013, pp. 152-158. doi: 10.5923/j.ijaf.20130304.05.
Article Outline
1. Introduction
- In recent decades, caffeine has been identified as a compound with allelopathic potential that causes the autotoxicity of coffee plants in older plantations since this purine alkaloid, mainly released by the fruits and leaves, accumulates in the soil over time. Caffeine accumulation is proposed to be one of the factors responsible for the observed worldwide premature decay phenomenon in unshaded older coffee monocultures[1, 2, 3]. However, several investigators have argued that soil microorganisms, such as Pseudomonas putida, Serratia marcescens, species of the genera Alcaligenes, Rhodococcus, and Klebsiella and several fungi mainly belonging to the Penicillium and Aspergillus genera and the yeast Trichosporon asahii, are able to degrade caffeine[4, 5]; some of these investigators, therefore, doubt the contribution of caffeine to the allelopathy of coffee[4]. Caffeine is often used as a specific marker for the detection of ground and surface water contamination with waste water, and several species of caffeine-degrading bacteria, such as Pseudomonas putida, Alcaligenes species and species of Klebsiella and Rhodococcus, have been isolated from domestic waste water or surface water[6]. Therefore, caffeine degradation may occur rather slowly in environments with a low density of suitable microorganisms. Moreover, caffeine is toxic to most soil microorganisms. Recently, a photo-Fenton process for caffeine degradation was developed as an alternative method to microbial degradation[7].While the biosynthesis of caffeine and related purine alkaloids has been extensively studied in Coffea arabica and Camellia ptilophylla and to some degree in Theobroma species, insights into the catabolism of caffeine in planta have been obtained solely by tracer experiments using 14C-labeled purine alkaloids[4]. Based on these data, a major catabolic pathway was constructed that starts with the demethylation of caffeine to produce theophylline and xanthine, which is subsequently degraded via the purine catabolic pathway. However, according to the MetaCyc database of metabolic pathways, the plant enzymes and genes responsible for the demethylation of the purine alkaloids, have not been identified and characterized[8]. In bacteria, demethylating and oxidative catabolic pathways are involved in the degradation of caffeine[9, 10,11]. In fungi, theophylline was described as an intermediate in the degradation pathway[12]. Roussos et al.[13] collected 272 strains of filamentous fungi collected from the soil and the coffee leaves and fruits in coffee-growing areas. Only 5 strains of Aspergillus species and 2 strains of Penicillium species, exclusively, were able to degrade the caffeine present in the culture medium. In another study, 131 epiphytic and endophytic fungi from coffee leaves were determined[13].Vega et al.[14] isolated hundreds of endophytes with an unknown role from coffee plants collected from different geographic sites. Thus, coffee plants harbor a high diversity of microorganisms which may be important for the prosperity of the particular ecosystem coffee plant-microorganisms. However, whether endophytes may drive caffeine degradation in planta, was not directed. The traditional coffee plantations in Coatepec (Mexico) have ground cover vegetation that is dominated by Poaceae and Asteraceae for sunny plots and Commelina species in the shade, sometimes in combination with trees[1, 2]. It may be speculated that in this ecosystem weeds and trees are able to reduce the amount of coffee allelochemicals in the soil by absorption of the compounds, but the contribution of the ground-cover plant species, intercropped species and their associated microorganisms in the reduction of caffeine in the soil is unknown.Aromatic plants, such as sage (Salvia officinalis), oregano (Origanum vulgare), mint (Mentha piperita) and basil (Ocimum basilicum), have been used as intercrops in coffee plantations. These aromatic plants stimulated the plagiotropic growth of the coffee plants, whereas their own growth was negatively influenced. In young plantations, the negative effect was reduced in basil and sage plants compared to other aromatic species[15, 16]. As found by uptake experiments performed in the IMBIO lab, University of in Bonn, all of the species were able to absorb caffeine, which accumulated in the roots. When basil was transferred to a caffeine-free medium, nearly all of the absorbed caffeine was released from the plant into the medium, whereas with sage, only a small portion could be detected. This result prompted us to elucidate the fate of the caffeine that is absorbed by sage, a species lacking purine alkaloids as natural constituents. In planta degradation of caffeine is of great importance when herbs like sage will be used as intercrops in coffee plantations. Presently no study is available that directs caffeine degradation in other plants than those belonging to the genera Coffea, Camellia and Theobroma.
2. Materials and Methods
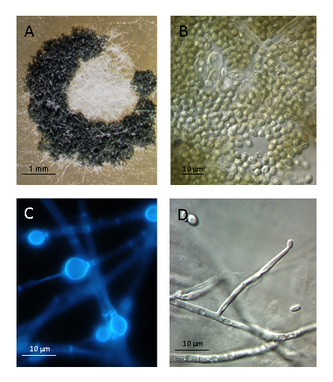
- Plant Material and Caffeine IncubationsSix-week-old Salvia officinalis plants (accession No. 23581), cultivated under greenhouse conditions, were incubated with 5 mM caffeine for 24 to 120 hours (h) in sterile tap water. In some experiments, the incubation medium was mixed with both Rifampicin (40 mg/mL) and Cefotaxime (250 mg/mL) (Duchefa Biochemie, The Netherlands) to suppress bacterial growth. Aliquots (100 µL each) of the incubation medium were stored at -20°C until analysis; the caffeine and degradation products were analyzed immediately after the thawing of the aliquots. For the determination of caffeine absorption, the roots and shoots of sage plants from all incubations were harvested separately and homogenized with quartz sand and 5 volumes (w/v) of 50% methanol. The homogenates were centrifuged, and the supernatants were analyzed for caffeine, theophylline and theobromine by HPLC using the method described in[14]. Caffeine and the degradation products, theobromine and theophylline, were identified by co-chromatography with synthetic compounds. For the experiments with sterile Salvia officinalis, the seeds were surface-sterilized with 80% ethanol for 1 min, followed by a 20 min treatment with 2% hypochlorite, supplemented with 0.1% Tween 20. After washing three times with sterile H2O, the seeds were transferred either to pots with sterilized soil (50% sand, 50% sieved compost; Hawita Kompost and sand, Kissener´s Gartenmarkt, Bonn, Germany) or cultivated in sterile glass jars with ½ strength Murashige and Skoog (MS) salts, plus vitamins and additionally containing 0.8% plant agar, 30 g/L sucrose and 0.5 g/L at a pH of 5.8. The plant cultivation was performed under sterile conditions in a Persival-Scientific culture chamber under conditions of 200C, 12 h of light (PAR: 100 microeinstein m-2 s-1), and 55% relative humidity. The plants were used for experiments after the development of 5 to 7 leaves. All experiments were repeated at least 3 times.Isolation of the root-associated microorganismsPieces of the sage roots were placed onto sterile Czapek or malt agar plates and incubated in the dark until the most predominant microorganisms were observed. Clean colonies of these plates and 200 ml of the Salvia officinalis incubation medium were used as the inoculum for cultures in sterilized, liquid Czapek or malt medium with and without the antibiotics, Rifampicin (200 µl of a 40 mg/mL stock solution) and Cefotaxime (200 µl of a 250 mg/mL stock solution). The inoculation flasks were incubated on a rotary shaker (100 rpm) at room temperature. After 3 days, the liquid culture was supplemented with caffeine (0.5 mM), and 200 µl from each flask was stored for the caffeine determination at the start of the incubation (day 0). The degradation of caffeine was monitored by HPLC over a period of 5 days by collecting and analyzing 200 µl samples of the media. At the end of the incubation period, the three microbial suspensions that were able to degrade caffeine were filtered with filter paper, and the supernatants were discarded after collecting 200 µl aliquots. Two fungi and one bacterial isolate capable of degrading caffeine (isolates ThSoW29508, RhSoW01 and PsSoW01) were placed on Czapek or malt agar and subcultured as described above. The isolates were each incubated in liquid medium (100 mL) with 5 mM caffeine, directly after the isolation and 8 weeks after the isolation.Identification of the microorganisms – barcoding PCR, cloning and sequencingTo characterize the isolate ThSoW29508 on a molecular level, mycelia grown on Czapek medium were scraped off the plates and ground in liquid nitrogen. DNA was extracted using the DNeasy plant mini kit (Qiagen, Hilden, Germany), according to the instructions of the manufacturer. Three primer combinations were chosen for the barcoding of the isolate ThSoW29508. For the PCR amplification of the Internal Transcribed Spacer Region (ITS), primers ITS1F (5´-TCCGTAGGTGAACCTGCGG-3´) and ITS4R (5´-TCCTCCGCTTATTGATATGC-3´), according to[17] were used. Additionally, a partial sequence of the large exon of tef1 was amplified with the primers tef1afw (5´-GTGAGCGTGGTATCACCATCG-3´) and tef1arew (5´-GCCATCCTTG GAGACCAGC-3´), and the primerpairChit42-1a (5´-GCTYTCCATCGGTGGCTG GAC-3`) and Chit42-2a (5`-GGAGTTGGGGTAGCTC AGC-3`) was used to amplify a fragment of the gene encoding endochitinase 42 (ech42)[18]. A second fungal isolate, RhSoWo01, cultivated on Sabouraud dextrose agar, was used for in-cell PCR with the primers ITS1F (5´-TCCGTAGGTGAACCT- GCGG-3´) and ITS4R (5´-TCCTCCGCTTATTGATATGC- -3´), according to[17]. The identification of the bacterial isolate, PsSoW01, cultivated on Pseudomonas agar F/P, was also accomplished by in-cell PCR with the primers P13P (5´-AGGCCCGGGAACGTATTCAC-3´) and P11P (5´-GAGGAAGGTGGGATGACGT-3´) for amplification of the 16S ribosomal RNA gene[19]. For all PCR reactions, either 20 ng of genomic DNA or cells were mixed with 5 µL Taq-buffer (10x), 5 µL MgCl2 (25 mM), 0.4 µL dNTPs (10 mM), 0.2 µL Taq-Polymerase (2.5 U, Fermentas) and 2.5 µL of each primer (10 pmol/µL) in a 50 µL PCR reaction. The parameters for the PCR reactions were as follows: 95°C for 5 min for the initial denaturation; 35 cycles each of 95°C for 30 seconds (sec), 58°C (primer pair ITS), 64°C (primer pair tef1), 63°C (primer pair Chit42) or 55°C (primer pair P13P/P11P) for 30 sec, respectively, 72°C for 1 min 30 sec; and a final extension at 72°C for 7 min. The PCR products were separated and visualized on a 1% agarose gel stained with ethidium bromide. The PCR products were purified with the Qiaquick PCR purification kit (Qiagen, Hilden, Germany) and used directly for sequencing. The PCR products were sequenced with the same primers used for the barcoding PCR using the Quick Start Kit (Beckman-Coulter, München, Germany). A magnetic bead purification (Agencourt Clean Seq, Beckman-Coulter, München, Germany) of the sequencing fragments was accomplished, and the fluorescent-labelled fragments were separated and detected at 6.0 kV for 60 min by means of capillary gel electrophoresis, equipped with a laser-induced fluorescent detector (CEQ 8800, Beckman-Coulter, München, Germany). The evaluation of the data and the alignments and the analysis of sequences were performed with Beckman-Coulter software, and the sequences were analysed by a Blast search in NCBI for the identification of the species.Fungal colonization of the plant tissueSterile plants were used for the colonization studies; plants were cultivated as described above. Inoculation of the plants was done by the application of a spore suspension (5 x 105 spores/mL) into the soil (50 mL/pot). The spores were harvested from fungal colonies grown on potato dextrose agar (PDA) for 14 days (d). After inoculation, seedlings were grown for 21 d in a controlled-environment chamber (16-h light/8-h dark photoperiod) at 25°C. For the re-isolation of Trichoderma hamatum, plants were harvested 21 days after inoculation by cutting the roots and stems into sections of 0.5 cm in length and up to a height of 5 cm above the soil line. Roots and stems were taken from 20 different plants. The plant samples were either non-sterilized or surface sterilized using NaOCl (1.3% available chlorine) for 1 min, followed by three washes in sterile, distilled water, and placed into Petri dishes on water agar containing antibiotics (50 mg/L penicillin, 50 mg/L chlortetracycline and 50 mg/L streptomycin). The plates were incubated at 24°C in the dark for 3 days and then transferred to the light (photoperiod of 12 h) for an additional 4 days. For the detection of fungal colonization, the samples were examined microscopically using a Leica MZ 16 F stereomicroscope (Leica, Bensheim, Germany) with 10-fold magnification. The numbers of samples with Trichoderma colonies on each Petri dish were recorded after 7 days, and the data were expressed as % isolation rate.Microscopic studiesInvestigations of fungal structures by light and fluorescence microscopy were carried out with a Leitz DMR photomicroscope (Leica, Bensheim, Germany). The fungal structures were stained in the dark with 0.05% Uvitex 2B (Syngenta, Basel, Switzerland) in 0.1 M Tris/HCl buffer, pH 8.0. The microscope was equipped with a digital camera (KYF 75 Sony) and Diskus software (TB Hilgers, Königswinter, Germany) for image processing.Statistical analysesValues were presented as means ± standard deviations from at least three independent treatments. These data were subjected to ANOVA analysis of variance (Bartlett´s test for homogeneity of variance, Tukey´s student range test and Dunn t test, P < 0.05) using SAS version 8.1.
3. Results
- Decrease of caffeine in the incubation mediumIn the incubation medium of non-sterile sage plants, the level of caffeine started to decrease to 80 to 90% after three days of incubation and dropped to 10 to 20% at day five (Fig. 1). Traces of theobromine could be found at days three and four. No other catabolite was detected. As for the sterile plants, the caffeine amount applied remained almost constant over five days. When the incubation medium of non-sterile plants was supplemented with the antibiotics Rifampicine and Cefotaxime, 75% of the caffeine was still present after five days.
|
 | Figure 4. Re-isolation of Trichoderma hamatum from surface-sterilized and non-sterilized roots and stem pieces of sterile-grown sage plants inoculated with Trichoderma spores |
4. Discussion
- While the demethylation reactions of caffeine were performed by the microorganisms, both the microorganisms and the sage plants could have been responsible for any further degradation. Because we could not detect any xanthine in the incubation medium, it might have been metabolized rapidly. Pseudomonas putida, a widespread saprotrophic soil bacterium, colonizes the roots of different plants. Strains of Pseudomonas species from the soil of coffee plantations, which showed positive chemotaxis toward caffeine, exhibit higher rates of caffeine degradation than other strains of this bacterial species[21]. Yu et al.[22] suggested that P. putida possesses multiple N-demethylases that are involved in caffeine and methylxanthine demethylation. R. glutinis was not yet known to degrade caffeine. To our knowledge this is a first report of Trichoderma hamatum being associated as an endophyte with Salvia officinalis and, furthermore, being involved in degrading caffeine in a Trichoderma -sage-interaction. The ability to degrade caffeine is of high importance when sage plants will be used as intercrops in coffee plantations.Trichoderma species, often found in the rhizosphere of plant roots, are widely used for the biocontrol of plant diseases and have been shown to alter plant metabolism by interacting with plant roots. These fungi can act as opportunistic, avirulent plant symbionts and induce systemic resistance against pathogens[23, 24, 25, 26]. However, cooperation with plants in degrading xenobiotics has not been described to date[27]. It has recently been demonstrated that Trichoderma hamatum exists as a beneficial endophyte in Theobroma cacao[28], and T. stromaticum is already known to exist endophytically in the vascular system of cocoa[29]. Numerous fungal endophytes have been isolated from coffee plants. Although not yet investigated[14], these plant-associated fungi may contribute to the degradation of caffeine in planta and, perhaps more generally, to the degradation of xenobiotic compounds, including allelochemicals. Here, we propose the existence of a beneficial biochemical interaction between endophytic Trichoderma hamatum and sage in caffeine degradation inside the root. It is likely that root-associated microorganisms have a more general role in caffeine degradation than was previously considered. Specifically, Trichoderma hamatum, acting in cooperation with sage plants by degrading the allelochemical caffeine, suggests a function of this fungus that has not yet been taken into account. The caffeine-induced growth inhibition, due to the high sensitivity of the root tips to this compound, might be explained by the not yet established colonization of young tissues by microorganisms and a lack of interactions with endophytic microorganisms. Depending on the dosage, volatile compounds of aromatic plants can have beneficial or harmful effects on neighbored plants. Low concentrations of some monoterpenes can promote plant growth whereas higher ones led to stomata opening and desiccation of receiver plants[15, 30]. A possible influence of caffeine on monoterpene synthesis in sage and how T. hamatum is affected by the sage monoterpenes or involved in the crosstalk is under investigation.
5. Conclusions
- In the agro-ecosystem “coffee plantation”, a high level of colonization of the soil with diverse, active microorganisms, along with suitable ground-cover vegetation, should contribute to a reduction of the deleterious effects caused by caffeine. Shaded coffee plantations are predestined for a better degradation of caffeine because of their high biodiversity[31] including a high richness in microorganisms and plant species[32]. Using sage with its endophyte Trichoderma hamatum as an intercrop may be a strategy to reduce caffeine accumulation in the soil. A possible role of the local ground cover vegetation in caffeine degradation with the aid of endophytes has still to be unraveled. A general function of endophytic microorganisms insupporting the plant´s efforts in the detoxification and degradation of xenobiotics has to be elucidated in future.
ACKNOWLEDGEMENTS
- The authors are thankful to Andreas Ulbrich for the support with the statistical analyses.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML