-
Paper Information
- Next Paper
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Agriculture and Forestry
p-ISSN: 2165-882X e-ISSN: 2165-8846
2012; 2(4): 150-156
doi: 10.5923/j.ijaf.20120204.03
Phylogenetic Relationships of Santalum album and its Adulterants as Inferred from Nuclear DNA Sequences
Anupama Chembath 1, Balasundaran M 1, Sujanapal P 2
1Forest Genetics and Biotechnology Division, Kerala Forest Research Institute, Peechi, Thrissur, Kerala, 680653, India
2Sustainable Forest Management Division, Kerala Forest Research Institute, Peechi, Thrissur, Kerala, 680653, India
Correspondence to: Balasundaran M , Forest Genetics and Biotechnology Division, Kerala Forest Research Institute, Peechi, Thrissur, Kerala, 680653, India.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
The East Indian sandalwood, Santalum album, valued for its fragrant oil yielding heartwood is a major ingredient in indigenous medicines and perfumes. Scarcity of sandal has led to illegal felling of sandal trees, and adulteration of sandalwood and oil. This study represents the first molecular phylogeny of S. album and its adulterant species Osyris wightiana, Erythroxylum monogynum, Buxus sempervirens, Ximenia americana, Osyris lanceolata, and Chukrasia tabularis through 18S and 26S rDNA sequencing. In the Maximum Parsimony (MP) tree for 18S and 26S rDNA data sets, moderate to high bootstrap support was obtained for the nodes. For 18S rDNA data sets, the tree had B. sempervirens and X. Americana as the upper branch, with E. monogynum branched separately to the cluster. The lower branch had S. album and O. wightiana with O. lanceolata joining separately to both clades of the tree. In the MP tree for 26S rDNA datasets, S. album and O. wightiana formed the major cluster with X. americana clustering separate and B. sempervirens and O. wightiana as the lower branch with C. tabularis clustering separate to the tree. The molecular data presented here provided useful information for resolving the phylogenetic relationship of these plants. Inferences from this study are in accordance with Cronquist’s system of classification of flowering plants where all the species originate from a single phylogenetic tree of Rosidae.
Keywords: Santalum Album, Adulterants, Gene Sequencing, Sequence Comparison, Molecular Phylogeny
Article Outline
1. Introduction
- Santalum album L., commercially known as East Indian sandalwood is a medium sized, xylem tapping, root hemi-parasitic tree belonging to the family Santalaceae. The species, commonly known as sandal is valued for its heartwood containing the precious sandal oil. The Indian sandalwood has the highest oil content (6 to 7%) and a desirable aroma profile, highly prized in perfumery and indigenous medicine[1].The annual production of sandalwood worldwide is estimated between 200 to 300 tonnes, of which 90 per cent is from India. Scarcity of sandal in open market and consequent price hike to exorbitant level have led to illegal felling and smuggling of trees. Sandal has been categorized as ‘Vulnerable’ in the Red Data List by IUCN[2]. Due to its commercial importance, sandalwood arriving for trade in the market is adulterated with many other indigenous as well as imported scented wood species.Rao et al.[3] have listed a few timbers that are used for adulterating sandalwood. Wood of S. album and Osyris species are strikingly similar in most of the wood anatomical characters.For this reason, sandalwood is often adulterated using Osyris spp. Osyris lanceolata Hochst. & Steud., a member of Santalaceae family, also called Tanzanian Sandalwood or East African Sandalwood, possesses scented heart wood. Trees of the genus Osyris are either shrubs or small evergreen trees, and are usually root hemi-parasites. But the oil, used in pharmaceutical and cosmetic industries abroad, lacks the sensuality of East Indian sandalwood oil.O. wightiana Wall. ex Wight. is found to occur rarely in higher altitudes (900 m above m.s.l.) of Idukki district of Kerala state[4] and in Tamil Nadu[5] in India. Erythroxylum monogynum Roxb., possessing fragrant heartwood, native to the Indian subcontinent is the source of ‘Indian bastard sandal’, also used to adulterate sandalwood[6]. Erythroxylaceae comprises 200 species distributed throughout the tropics. The root bark contains alkaloids; the heartwood is light brown which is very durable and easy to work.Other scented woods often used to adulterate sandalwood are Buxus sempervirens L., Ximenia americana L. and Chukrasia tabularis var. velutina. B. sempervirens, commonly known as American boxwood, a member of the Buxaceae family is an evergreen shrub native to Western and Southern Europe, Northwest Africa and Southwest Asia[7]. Wood is very hard and heavy, used for engraving, marquetry and wood turning. X. Americana, belonging to Olacaceae family, also known as ‘false sandalwood’ is a small, shrubby tree native to Central and South Florida and the African tropics[8]. It has hemi-parasitic roots, but it does not require a host to thrive. Bark and roots are used for tanning and wood for firewood and charcoal. C. tabularis, a member of the Meliaceae family, locally known as ‘agil’ in Kerala, is a deciduous medium-sized tree found in India, Bangladesh, China, Thailand and Malaysia[9]. Freshly cut wood has a fragrant odour, but dried wood has no characteristic odour. Planed surfaces have a high lustrous satiny sheen. The timber is highly prized for superior cabinet work, decorative panelling, interior joinery, carving, toys and turnery.The state of knowledge about relationships among the various lineages of land plants is currently incomplete[10]. Clarification of phylogenetic relationships among Santalales and their adulterant groups presents opportunities to better understand their evolutionary and interfamilial relationships. Ribosomal RNA or DNA sequences (rRNA/ rDNA) have frequently been used to reconstruct deep branches of evolutionary history. The gene containing highly conserved as well as variable regions facilitate alignments of nucleotide sequences derived from phylogenetically linked taxa[11]. 18S rRNA/rDNA has been used for phylogeny reconstruction within many groups of eukaryotes[12]. Although phylogenetic analysis of 18S rDNA sequences provides critically independent data set for the assessment of higher-level relationships, in many instances analysis of 18S rDNA sequence alone will not provide adequate resolution. Combining 26S rDNA sequence data and 18S rDNA sequence data would increase the quantum of phylogenetically informative history fourfold and provide greater resolution and support at higher taxonomic levels [13]. The present investigation has been designed to deduce the evolutionary relationships among S. album and its adulterant wood species using rDNA sequencing and comparison studies.
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
- Semi-dry logs and leaf samples of Santalum album were collected from Marayur Sandalwood reserve forest located on the leeward side of the Western Ghats in the South West region of India. Samples of Osyris wightiana and Erythroxylum monogynum were collected from Chinnar wildlife sanctuary located adjacent to the Marayur sandal reserve forest. All these places lie at 10º15’ N latitude and 77º11’ E longitude. DNA extractions were accomplished using QIAGEN DNeasy Plant Mini Kit and the DNA was used as template for PCR amplification of ribosomal DNA using specific primers designed and developed for Santalum and Osyris species.
2.2. Primer Design and Synthesis
- From the 18S and 26S ribosomal RNA partial sequences of S. album (L24416, AY957453) and Osyris lanceolata (U42803, AF389274) deposited in the NCBI nucleotide library, specific primers capable of amplifying partial ribosomal DNA units were designed [14] (Table 1). The primers were synthesized at MWG Biotech Pvt. Ltd., Bangalore, India. Using these primers, we PCR amplified 18S and 26S rDNA of S. album and O. wightiana using DNA from leaf samples of the species collected from Marayur as template. The primer sets used for amplifying O. wightiana loci were used for the amplification of loci from E. monogynum, collected from the same area.
2.3. PCR Amplification and Sequencing
- PCR amplification reactions were performed using FINNZYMES High Fidelity PCR Kit. The PCR products were subjected to sequencing at MWG Biotech Pvt. Ltd. The 26S rDNA region of E. monogynum could not be amplified; hence it was not included in the present study. The sequencing was performed using BigDye terminator v3.1 cycle sequencing Kit containing AmpliTac DNA polymerase (Applied Biosystems).
2.4. Phylogenetic Analyses
- Phylogenetic analyses were conducted using Molecular Evolutionary Genetics Analysis (MEGA) software version 4[15]. Sequences were manually aligned using Alignment Explorer (AE) in MEGA 4 and separate alignments were created for 18S and 26S rDNA data sets. Sequences of 18S and 26S rDNA of sandalwood adulterants Buxus sempervirens (L54065, AF389243), Ximenia americana (L24428, DQ790220) and Osyris lanceolata (U42803, AF389274), and 26S rDNA of Chukrasia tabularis (AY128154) deposited in the NCBI nucleotide library were chosen as outgroups and were included in the alignment. The 18S rDNA sequence of C. tabularis was unavailable in the NCBI nucleotide library; hence it was not included for the analysis. The evolutionary history was inferred using the Maximum Parsimony (MP) method[16]. The consistency index, retention index and the composite index for all sites and parsimony-informative sites were calculated. The MP tree was obtained using the Close-Neighbour-Interchange algorithm[17] with search level 2 in which the initial trees were obtained with the random addition of sequences (100 replicates). All positions containing gaps and missing data were eliminated from the dataset using complete deletion option. Nucleotide pair frequencies including identical, transitional and transversional pairs and the overall transition/transversion bias were estimated and substitution patterns were calculated. Tajima’s relative rate test for testing molecular clock hypothesis was performed to test the constancy of evolutionary rates between two sequences or clusters of sequences, using an outgroup sequence. P-value and χ2 test statistics were calculated with one degree of freedom[18].In the bootstrap test of phylogeny using Maximum Parsimony method, the consensus tree inferred from 1000 replicates were taken to represent the evolutionary history of the taxa analyzed[19]. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) was shown next to the branches of the out tree.
2.5. Sequence Comparison and Similarity Search
- Comparison of 18S and 26S rDNA sequences of S. album, O. wightiana and E. monogynum with outgroup sequences of B. sempervirens, X. americana, O. lanceolata and C. tabularis were achieved through the bioinformatics tool for multiple sequence alignment, CLUSTAL W (1.83). Sequence similarity search for 18S and 26S rDNA sequences of S. album, O. wightiana and E. monogynum were accomplished by means of NCBI-BLAST.
3. Results
- Aligned datasets for 18S rDNA genes contained 1554 positions, out of which 1198 were parsimony informative. The consistency index was 0.7480, the retention index 0.3735, and the composite index 0.2975 for all sites and parsimony-informative sites. Likewise, for 26S rDNA dataset there were a total of 740 positions in the final dataset, out of which 540 were parsimony informative. The consistency index was 0.7827, the retention index 0.5135, and the composite index was 0.4278 for all sites and parsimony informative sites. Nucleotide pair frequency estimations in 18S and 26S rDNA sequences, results of substitution pattern calculations with S. album as the reference sequence, and the overall transition/transversion bias are provided in Table 2.Tajima’s relative rate test for testing molecular clock hypothesis in 18S and 26S rDNA sequences was performed with S. album (lineage 1) kept constant (Table 3).In the bootstrap test of phylogeny using Maximum Parsimony (MP) method the bootstrap consensus tree inferred from 1000 replicates was taken to represent the evolutionary history of the taxa analysed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test is shown next to the branches of the out tree. In the MP tree for 18S rDNA data sets (Figure 1) the most parsimonious tree was with length 4272. Moderate to high bootstrap support was obtained for the nodes, and the lower branch formed a clade (A) of Santalales with S. album and O. wightiana (75% BS) clustered together. The upper branch had high BS support (100%) for B. sempervirens and X. Americana (Clade B), and also had E. monogynum separately branched to that cluster, but with low BS support (<50%). O. lanceolata joined separately to both the clades of the tree.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
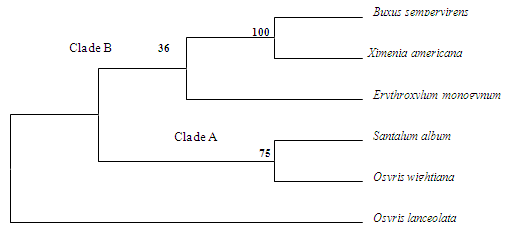 | Figure 1. Bootstrap consensus tree (MP) of 18S rDNA dataset. Numbers above nodes are bootstrap percentages (1000 replicates) |
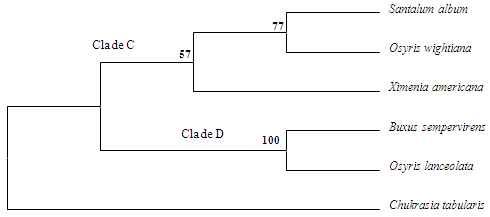 | Figure 2. Bootstrap consensus tree (MP) of 26S rDNA dataset. Numbers above nodes are bootstrap percentages (1000 replicates) |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
- In the MP tree for 26S rDNA datasets (Figure 2) the most parsimonious tree was with length 1840. MP resolved a monophyletic Santalales (Clade C) consisting of S. album and O. wightiana (BS 77%) in the major cluster with X. americana clustering separate (BS 57%). The next clade (D) had high BS support (100%) for B. sempervirens and O. wightiana. C. tabularis clustered separate to the 26S MP tree.Comparisons between 18S and 26S rDNA sequences and their outgroups using bioinformatics tool CLUSTAL W (1.83) for multiple sequence alignment gave scores indicative of the identities and differences between the sequences (Table 4). In the 18S sequence scores the most identical species towards S. album were O. wightiana and O. lanceolata giving a score of 98, and the least identical being B. sempervirens with a score of 94. At the same time, for 26S rDNA sequence scores also, the most identical scores were for O. wightiana and O. lanceolata with 92, while the least one being C. tabularis with 62 as the identity score.
4. Discussion
- This study represents the first molecular phylogeny of S. album and its adulterant species inferred from 18S and 26S rDNA sequences. The molecular data presented here, though limited in number, provides novel information useful in resolving the phylogenetic history of these plants. By following Bentham and Hooker[20] with recent circumscription of certain families to their current concept[21], all the species selected for the study belong to the class Dicotyledonae of natural system. In all the cases, the floral lobes are 3-6 with perianth, with or without calyx, and corolla lobes which are free or connate. Principal species, S. album and its closest natural relatives O. wightiana and O. lanceolata belong to the family Santalaceae under the order Santalales belonging to the subclass Monochlamydeae. B. sempervirens belonging to the family Buxaceae is the next advanced relative in the natural hierarchy. In the natural system of classification, Santalaceae and Balanophoraceae of the order Unisexuales in Monochlamydeae are immediately followed by Buxaceae and Euphorbiaceae.In the 18S MP tree, monophyletic Clade A of Santalales consisted of S. album, O. wightiana (75% BS) clustered together. A polyphyletic Clade B composed of B. sempervirens and X. americana (100% BS) and E. monogynum separately branched to that cluster (BS <50%). O. lanceolata branched separately and joined to both the clades of the 18S MP tree. Among the species selected for the study S. album, O. wightiana, O. lanceolata and B. sempervirens are considered as advanced amongst Dicotyledonae. Species such as E. monogynum and X. americana are in the primary primitive group in hierarchy of class Dicotyledonae. Hence, these species are broadly categorized into two clusters- one advanced and the other primitive.In the 26S MP tree, a monophyletic Santalales (Clade C) was resolved comprising S. album and O. wightiana (BS 77%) with X. americana clustering separately (BS 57%). Clade D was found to be polyphyletic and had high BS support (100%) for B. sempervirens and O. lanceolata. C. tabularis clustered separate and was connected to both the clades of the 26S MP tree. In the phylogenetic system of classification,[22] all the species belong to the division Magnoliophyta and class Magnoliopsida. Except B. sempervirens all other species belong to the subclass Rosidae of phylogenetic tree. B. sempervirens belongs to the subclass Hamamelididae which is primitive than the subclass Dillenidae and Rosidae in the phylogenetic hierarchy and is the most primitive among all other species selected for the study. Among the Rosidae members, S. album, O. wightiana, O. lanceolata and X. americana belong to the order Santalales; within them the closest being Santalaceae and Olacaceae. Olacaceae is considered as the most primitive among Santalalean families as it comprises both root parasitic and nonparasitic species. Kuijt[23] considered the Olacaceae the Plexus from which the other Santalalean families were derived. E. monogynum the immediate relative of Santalanae group belonged to the Linales under Geraninae and is the most primitive of all the above species. While considering all the species selected for the study C. tabularis of the order Rutales of Rutanae group is the primitive among the Rosidae members. Study of phylogenetic relationships of Santalales and their relatives by Nickrent and Franchina using 18S and 26S rDNA sequences indicated that sequence of Buxus when compared with several genera of Rosidae, the genus nested within the clade composed of Cornales and Apiales and not in a basal position near or outside Fabales. Analyses by Nickrent et al. have placed Ximenia in the clade D of Olacaceae suggesting that haustorial parasitism arose just once in Santalales. In their study, Santalum and Osyris clustered together forming a well supported clade (clade E) of Santalaceae. The molecular evidences presented in a study by Der and Nickrent[24] corroborated the polyphyletic nature of Santaleae and illustrated Santalum being strongly supported clade, but relationships among its various subclades sometimes being poorly resolved. Phylogenetic relationships among early-diverging eudicots based on four genes grouped Osyris into core eudicots and Buxus into early diverging eudicots[25].A revised classification of Santalales was presented by Nickrent et al[26] in which the molecular and morphological data on santalalean clades were drawn together and a revised classification for the entire order was proposed. They came across several instances in which support values along the portions of the molecular tree were low (polytomies), thus not providing full resolution of interfamilial relationships. They also suggest that the tendency for monogeneric groups to occur as sister to species-rich clades is not unusual within angiosperms, and in such situations, circumscribing monogeneric families appears to be the best solution.Angiosperm phylogeny by Soltis et al[27] revealed that relationships among major clades of Santalales have not been resolved with strong BS support in recent analyses, and many segregate families have been newly recognized.An angiosperm phylogeny was reconstructed using four slowly evolving mitochondrial genes by Qiu et al[28] in which S. album and X. americana clustered together within Santalales, which is a member of an expanded asterid clade. B. sempervirens clustered within Buxales which forms a basal grade that diverged before the diversification of asterids. Phylogenetics of early branching eudicots were examined by Barniske et al[29] which also placed Buxales as successive sisters to core eudicots. In a study by Moore et al[30] which looked at the phylogenetic analysis of plastid genes, Maximum-likelihood analyses of the gene alignment placed Santalales as successive sisters to Asteridae, with each node receiving 99-100% Bootstrap support.However, in Cronquist system[31], all the species originate from a single phylogenetic tree of Rosidae and Chukrasia tabularis is the distant relative. Earlier workers recognized the polyphyletic relationship of subclass Rosidae and its orders such as Santalales. These selected diverse group of species showed different mode of nutrition and habitat not only owing to its phylogeny, but also due to adaptation or parallel evolution. Results of the present study are in agreement with the Cronquist classification and the cladistic data provided more insight into the phylogenetic relationships of these biochemically related taxa in several aspects. Though, a wider sampling and information on sequences of other genes will be required to fully resolve the relationships in Santalales, 18S and 26S rRNA sequences are informative on recent divergences and relationships within major groups of land plants.
5. Conclusions
- The East Indian sandalwood, Santalum album, valued for its fragrant heartwood containing sandal oil is a major ingredient in cosmetics, indigenous medicines and perfumes. Scarcity of sandal for free trade and consequent exorbitant price has led to illegal felling of sandal trees, thereby causing further depletion of its natural resources, and adulteration of sandalwood and oil. Elucidation of phylogenetic relationships among Santalales and their adulterant group presents opportunities to understand their evolutionary and interfamilial relationships. In this study, we illustrate the phylogenetic analyses of Santalaceae and its adulterant wood species using rDNA sequencing and comparison studies. Using specific primers, 18S and 26S ribosomal DNA of Santalum album, Osyris wightiana and Erythroxylum monogynum were PCR amplified and the genes were sequenced. Sequence comparisons together with out-group sequences of Buxus sempervirens, Ximenia americana, Osyris lanceolata, and Chukrasia tabularis were achieved through CLUSTAL W and sequence similarity search using NCBI-BLAST. Phylogenetic analyses were done using Molecular Evolutionary Genetics Analysis (MEGA) software. In the Maximum Parsimony (MP) tree for 18S and 26S rDNA data sets, moderate to high bootstrap support was obtained for the nodes. While considering all the species selected for the study, C. tabularis of the order Rutales is the primitive among the Rosidae members. Inferences from the present study are in accordance with the Cronquist’s classification of flowering plants (1988) where all the species originate from a single phylogenetic tree of Rosidae and Chukrasia tabularis is the distant relative. Cladistic data provided more insight into the phylogenetic relationships of these biochemically related taxa in several aspects.
ACKNOWLEDGEMENTS
- We are grateful to the Kerala Forest Department for permission to collect the samples. The study was funded by Kerala State Council for Science, Technology and Environment.
References
| [1] | Sanjaya, Ananthapadmanabha H S, Ravishankar Rai, V, “In vitro shoot multiplication from the mature tree of Santalum album L.” in Sandal and Its Products, ACIAR Proceedings No. 84. ACIAR, Canberra, Australia, pp. 60-65, 1998. |
| [2] | IUCN., IUCN Red List of Threatened Species. Version 2009.1, www.iucnredlist.org, 2009. |
| [3] | Rao R V, Hemavathi T R, Sujatha M, Chauhan L, Raturi R, “Stemwood and rootwood anatomy of Santalum album L. and the problem of wood adulteration”, in Sandal and Its Products, ACIAR Proceedings No. 84. ACIAR, Canberra, Australia, pp. 93-102, 1998. |
| [4] | Sasidharan N, “Study on the flora of Chinnar Wildlife Sanctury”, KFRI Research Report No. 167. Kerala Forest Research Institute, Peechi, Trichur, Kerala, India, pp. 24-62, 1999. |
| [5] | Mathew K M, “Materials for a flora of the Tamil Nadu Carnatic”, The Ranipet Herbarium, St. Joseph’s college, Tiruchirapalli, India, pp. 326, 1981. |
| [6] | Pullaiah T, Prabhakar C, Raviprasad R B, http://www.ars-grin.gov/cgi-bin/npgs/html/taxon.pl?414291, 1998. |
| [7] | Gilman E F, “Buxus sempervirens - Fact Sheet FPS-80”, Environmental Horticulture Department, Florida Cooperative Extension Service, Institute of Food and Agricultural Sciences, University of Florida, http://edis.ifas.ufl.edu, 1999. |
| [8] | Adewunmi C O, Sofowora E A, “Preliminary screening of some plants extracts for molluscicidal activity”, Planta Medica, vol.39, pp. 57-65, 1980. |
| [9] | Mabberley D J, Pannell C M, Sing A M, http://www.hear.org/Pier/species/chukrasia_tabularis.htm, 1995. |
| [10] | Nickrent D L, Malécot V, “A Molecular Phylogeny of Santalales”, in Fer A, Thalouarn P, Joel D M, Musselman L J, Parker C, Verkleij J A C, eds. Proceedings of the 7th International Parasitic Weed Symposium, Nantes, France, pp. 69-74, 2001. |
| [11] | Nickrent D L, Franchina C R, “Phylogenetic relationships of the Santalales and relatives”, Journal of Molecular Evolution., vol 31, pp. 294-301, 1990. |
| [12] | Soltis D E, Soltis P S, Nickrent D L, Johnson L A, Hahn W J, Hoot S B, Sweere J A, Kuzoff R K, Kron K A, Chase M W, Swensen S M, Zimmer E A, Chaw S. M, Gillespie L J, Kress W J, Sytsma K J, “Angiosperm phylogeny inferred from 18S ribosomal DNA sequences”,Annals of the Missouri Botanical Garden, vol 84,pp. 1–49, 1997. |
| [13] | Kuzoff R K, Sweere J A, Soltis D E, Soltis P S, Zimmer E A, “The phylogenetic potential of entire 26S rDNA sequences in plants”, Molecular Biology and Evolution, vol 15, pp. 251–263, 1998. |
| [14] | Anupama C, Balasundaran M, “Molecular detection of sandalwood adulteration”, in Conservation, Improvement, Cultivation and Management of sandal (Santalum album L.), Proceedings of National seminar, Institute of Wood Science and Technology (ICFRE), Bangalore, pp. 172-178, 2008. |
| [15] | Tamura K, Dudley J, Nei M, Kumar S, “MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0”, Molecular Biology and Evolution, vol 24, pp.1596-1599, 2007. |
| [16] | Eck RV, Dayhoff M O, “Atlas of Protein Sequence and Structure”, National Biomedical Research Foundation, Silver Springs, Maryland, USA, 1966. |
| [17] | Nei M, Kumar S, “Molecular Evolution and Phylogenetics”, Oxford University Press, New York, pp. 333, 2000. |
| [18] | Tajima F, “Simple methods for testing molecular clock hypothesis”, Genetics, vol 135, pp.599-607, 1993. |
| [19] | Felsenstein J, “Confidence limits on phylogenies: An approach using the bootstrap”, Evolution, vol 39, pp. 783-791, 1985. |
| [20] | Bentham G, Hooker J D, “Genera Plantarum” L. Reeve and Co, London, 1862-1883. |
| [21] | Brummitt KR, Powell C E, “Authors of Plant Names”, Royal Botanic Gardens, Kew, 1992. |
| [22] | Takhtajan A, “Diversity and Classification of Flowering Plants”, Columbia University Press, New York, pp. 643, 1997. |
| [23] | Kuijt J, “Mutual affinities of Santalalean families”, Brittonia, vol 20, pp.136– 147, 1968. |
| [24] | Der P J, Nickrent D L, “A molecular phylogeny of Santalaceae (Santalales)”, Systematic Botany, vol 33, pp. 107-116, 2008. |
| [25] | Kim S, Soltis D E, Soltis P S, Zanis M J, Suh Y, “Phylogenetic relationships among early-diverging eudicots based on four genes: were the eudicots ancestrally woody?”,Molecular Phylogenetics and Evolution, vol 31, pp.16–30, 2004. |
| [26] | Nickrent D L, Malecot V, Vidal-Russell R, Der P J, “A revised classification of Santalales”, Taxon, vol 59(2), pp. 538-558, 2010. |
| [27] | Soltis D E, Smith S A, Cellinese N, Wurdack K J, Tank D C, Brockington S F, Rrfulio-Rodriguez, Walker J B, Moore M J, Carlsward B S, Bell C D, Latvis M, Crawley S, Black C, Diouf D, Xi Z, Rushworth C A, Gitzendanner M A, Sytsma K J, Qiu Y L, Hilu K W, Davis C C, Sanderson M J, Beaman R S, Olmstead R G, Judd W S, Donoghue M J, Soltis P S, “Angiosperm phylogeny: 17 Genes, 640 Taxa”, American Journal of Botany, vol 98(4), pp.704-730, 2011. |
| [28] | Qiu Y L, Li L, Wang B, Xue J Y, Hendry T A, Li R Q, Brown J W, Liu Y, Hudson G T, Chen Z D, “Angiosperm phylogeny inferred from sequences of four mitochondrial genes”, Journal of systematics and evolution, vol 48(6), pp.391-425, 2010. |
| [29] | Barniske A M, Borsch T, Muller K, Krug M, Worberg A, Neinhuis C, Quandt D, “Phylogenetics of early branching eudicots: Comparing phylogenetic signal across plastid introns, spacers, and genes”, Journal of systematics and evolution, vol 50(2), pp. 85-108, 2012. |
| [30] | Moore M J, Soltis P S, Bell C D, Burleigh J G, Soltis D E, “Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots”, Proceedings of the National Academy of Sciences, vol 107(10), pp 4623-4628, 2010. |
| [31] | Cronquist A, “The Evolution and Classification of Flowering Plants” (Second edition), The New York Botanical Garden, New York, p. 643, 1988. |
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-Text HTML
Full-Text HTML