-
Paper Information
- Next Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Journal of Health Science
p-ISSN: 2166-5966 e-ISSN: 2166-5990
2014; 4(2): 25-33
doi:10.5923/j.health.20140402.01
Genetic Diversity of Breast Cancer in Senegalese Women: New Insight from Somatic Mutations
Fatimata Mbaye1, 2, Ahmadou Dem3, Malick Fall1, Gora Diop1, Babacar Mbengue4, Rokhaya N. Diallo5, Maguette S. Niang4, Mamadou Kane2, Sidy Ka3, Alioune Dieye4, Mbacké Sembene1, 2
1Département de Biologie Animale, Faculté des Sciences et Techniques, Université C.A., Diop, B.P. 5005 Dakar, Senegal
2Centre de Biologie pour la Gestion des Populations, Institut de Recherche pour le développement, IRD/Bel-Air, Senegal
3Institut du cancer, Faculté de Médecine, Pharmacie et d’Odonto-Stomatologie, Université Cheikh Anta Diop, Dakar Sénégal
4Institut Pasteur-Avenue Pasteur B.P.220-Dakar Sénégal
5Laboratoire de Biochimie Pharmaceutique, Faculté de Médecine, Pharmacie et d’Odonto-Stomatologie, Université Cheikh Anta Diop, Dakar Sénégal
Correspondence to: Fatimata Mbaye, Département de Biologie Animale, Faculté des Sciences et Techniques, Université C.A., Diop, B.P. 5005 Dakar, Senegal.
| Email: |  |
Copyright © 2014 Scientific & Academic Publishing. All Rights Reserved.
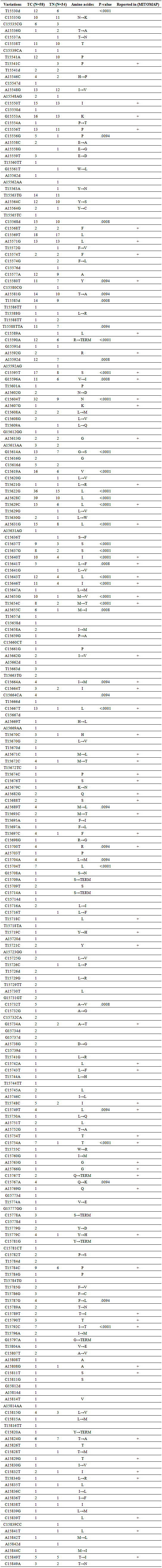
The purpose of this study is to determine the distribution of the mutations in MTCYB breast cancer and Genetic diversity of tumors. We sequenced MTCYB in the Senegalese patients suffering from breast cancer (N=58) associated with normal tissues (N=34). The sequences MTCYB of cancer and normal tissues were compared to the sequence revised Cambridge in MITOMAP database. Genetic diversity of tumors was studies using MEGA.6 and DnaSP 5.10 software. 96.55% (56/58) of tumors have somatic mutations. In our series 237 variations of MTCYB including 67 described previously in the MITOMAP database were noted. The frequency of mutations MTCYB tumors (90.29%) is higher compared with normal tissues (41.77%). 40 variations including 22 described as new show significant differences (P <0.05) between cancer tissues and normal tissues. Changes induced in a 58.28% amino acid change. A correlation between haplogroup U (P<0.0001) and the incidence of breast cancer was noted in the Senegalese population. dN> dS, MTCYB is involved into the carcinogenesis by accumulating non-synonymous mutations. Our results indicate that alterations MTCYB are common in breast cancer.
Keywords: Cancer, Breast, Somatic mutations, DNAmt, MTCYB
Cite this paper: Fatimata Mbaye, Ahmadou Dem, Malick Fall, Gora Diop, Babacar Mbengue, Rokhaya N. Diallo, Maguette S. Niang, Mamadou Kane, Sidy Ka, Alioune Dieye, Mbacké Sembene, Genetic Diversity of Breast Cancer in Senegalese Women: New Insight from Somatic Mutations, Journal of Health Science, Vol. 4 No. 2, 2014, pp. 25-33. doi: 10.5923/j.health.20140402.01.
Article Outline
1. Introduction
- Mitochondrial alterations associated with cancer have been identified and described in the literature. Such alterations include changes of mitochondrial gene expression [1; 2], mitochondrial structural abnormalities, or abnormal quantitative enzymatic components of the respiratory chain [3] and, more recently, through the development of molecular biology techniques, mutations of mtDNA. It is now well established that germline mtDNA mutations play an important role in diseases such as Leber hereditary optic neuropathy or Leigh syndrome [4]. However, unlike these diseases are somatic mtDNA mutations that are observed in cancers that are solid tumors [5] or hematologics [6]. Fraction of mtDNA mutations identified in human tumor cells is considered to have significant effects on mitochondrial function [7]. In summary, mtDNA mutations causing mitochondrial dysfunction may be the cause of tumorigenesis. Somatic mutations of mtDNA have been reported in various tumor types, including breast, colorectal, bladder, stomach, esophagus, lung, mouth, head tumor and neck and leukemia. These mtDNA mutations include point mutations, deletions and insertions [8; 9]. The roles that these changes play of mtDNA in the tumorigenesis and the development of the tumor have not been determined.Cytochrome B (MTCYB) is a region of the mitochondrial genome, located in positions 14747 and 15887 [10]. The MTCYB is the only gene of the mtDNA encoded respiratory complex III subunit. The MTCYB is a component of the respiratory chain also known as the bc1 complex or c reductase ubiquinol-cytochrome complex III. It is involved in the binding of substrate to quinone and is responsible for the transfer of electrons by which the transmembrane redox energy is converted into a proton motive power. In other words, the complex III plays a key role in the cells [11].In this study we determined the frequency and distribution of the mutations in MTCYB breast cancer and whether there is a correlation between mtDNA haplogroups and breast cancer.
2. Material and Methods
2.1. Samples
- After receiving the approval of the ethics committee of the University Cheikh Anta Diop of Dakar, informed consent, written in a standardized form was obtained from patients who have been the subject of this study. Tumor samples from patients with breast cancer (N = 58) and normal tissues that serve as controls (N = 34) were collected from a surgery performed at the Institute of Cancer Aristide Hospital Le Dantec. Samples were stored in 96° alcohol and frozen until use.
2.2. DNA Extraction, Amplification and Sequencing
- Whole genomic DNA was extracted from tissues after digestion with proteinase K and purified on a column (QIAGEN) as previously described [12]. The Cytochrome b gene was amplified by PCR using two primers, H15915 (TCT-CCA-TTT-CTG-GTT-TAC-AAG-AC) and L14723 (ACC-AAT-GAC-ATG-AAA-AAT-CAT-GGT-T). The PCR reactions were carried out in an Eppendhorf thermal cycler with an initial denaturation step at 94°C for 3 min. followed by 40 cycles corresponding to 45 sec. of denaturation at 92°C, 1 min. annealing at 50°C, and 1 min. 30 sec. elongation at 72°C. A 10 min. elongation step was performed after the final cycle. The obtained amplicons were shipped to Macrogen, South Korea for purification and sequencing using primer H15915.
2.3. Genetic Analyses
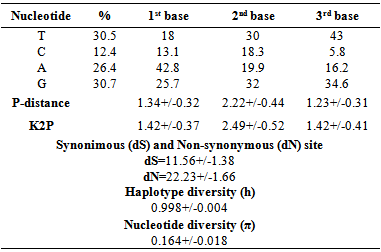
- The sequences obtained from normal and cancerous tissues are carefully checked, adjusted and aligned with BioEdit Version 7.1.9 editor [13] using the Clustal W algorithm [14]. The sequences MYCYB of normal and cancerous tissues were compared to the sequence revised Cambridge (NC_012920) [15] in MITOMAP database (www.mitomap.org). In the same database, each submitted sequence is assigned to a haplogroup. Somatic mutations that are not found in MITOMAP database are considered as new.Genetically, understanding polymorphism DNA sequence following nucleotide substitution is of primary interest. MEGA 6 [16] was used to calculate the nucleotide frequency, the proportion of nucleotide differences per site using the nucleotide p- distance model and the Kimura 2-p distance (K2P) model and the average number of synonymous substitution (S) and nonsynonymous substitution (N) per site using the Nei and Gojobori model [17]. The nucleotide frequency Nucleotide p- distance and K2P were calculated for the 1st, 2nd and 3rd base of each codon. Differences of synonymous and non-synonymous substitution were calculated between the sequences taken in pairs. Two standard indices of genetic diversity were estimated using the DnaSP program Version 5.10.01 [18]. These were the haplotype diversity (h) and the nucleotide diversity (π).
2.4. Analyses Statistiques
- Statistical analysis was performed using the results of percentage of mutants. Statistical analyzes comparing quantitative variables were performed by χ2 test with Statview software. For all statistical analyzes the threshold of 5% was chosen as significant.
3. Results
- Compared to the reference sequence of Cambridge (rCRS) 237 variations MTCYB were detected at the 67 which were already reported in the MITOMAP database (Table 1). Two (2) tumors presented no change, three (3) tumors had one (1) mutation and others (53) had multiple mutations with a total of 237 mutations. A total, 40 variations including 22 described as new show significant differences (P <0.05) between cancer tissues and normal tissues (Table 1). A frequency variation is very high in cancer tissues (90.29%) compared to normal tissues (41.77%). The changes consist of deletions, insertions and substitutions with 12.65%, 13.08% and 73.83% respectively.
|
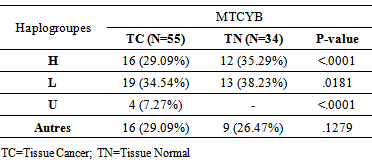
- Haplogroups were assigned to one of three major haplogroups of the African population (H, L and U) with a frequency> 10% [19]. Samples that could not be clearly attributed to a haplogroup were excluded. Three haplogroups H, L and U are significant differences between cancer tissues and normal tissues (P <0.0001; P = 0.181 and P <0.0001). Haplogroups are listed in Table 2.
|
|
4. Discussion
- In this study, we present a thorough analysis of MTCYB somatic mutations. Two (2) tumors presented no change, three (3) tumors had one (1) mutation and others (53) had multiple mutations with a total of 237 mutations. 67 mutations have been reported in other types of pathologies (MITOMAP) and 170 have been reported for the first time. Multiple mtDNA mutations are common in human tumors. This high mutation rate is called hyper-mutagenesis mtDNA, probably via the cellular oxidative stress [20]. 73.83% changes are made substitutions to 58.28% inducing a change in amino acid. Biochemical studies are needed to determine the effect of these mutations on the energy metabolism in the tumor cell. The rate of mutations in cancer tissues MTCYB is 90.29%. Genetic mutations MTCYB were reported in tumors of different anatomical origins, but the functional significance of these mutations is not well studied. A total of 40 variations including 22 described as new, are significantly different (P <0.05) between healthy tissue and cancerous tissue. Interesting fact is the absence of changes C15664CA; A15689T; C15700T; C15704A; C15704T; C15732T; C15749T; C15767A; T15787G and T15792C in normal tissues with a significant difference (P <0.05) compared to tumor tissue.To determine whether mtDNA haplogroups contribute to the development of breast cancer in the Senegalese population, we conducted a case-control study. These haplogroups were identified by polymorphism MTCYB in the Senegalese population. Comparing haplogroups of this study with that of the African population [19], the% mitochondrial haplogroup H is increasing and low for haplogroups L and U. Interesting fact is the absence of haplogroup U in normal tissues with a significant difference compared to cancerous tissues. In the Senegalese population haplogroup U could have an influence on the incidence of breast cancer. An association of haplogroup U with prostate cancer and kidney was noted by Booker et al [21]. The legacy of the mitochondrial haplogroup U is associated with an increased risk of prostate cancer (~ 2 times) and kidney cancer (2.5 times higher) among North American individuals. Therefore people with this haplotype are in a high risk group. The mitochondrial haplogroup U is found in 9.35% of the white population of the United States, there are over 20 million people in this high-risk group [21]. Haplogroup U is in 15% of the African population [19]. One limitation of our study is the sample size. The determination of different haplotype groups present in the Senegalese population is necessary to correlate with the incidence of breast cancer. Clinical parameters, metastases; hormone receptors must be correlated with the haplogroups a larger sample.Beckman & Ames have studied the general characteristics of mtDNA mutations [22]. They found that the majority of mutations were transitions C-T and A-G. What are correlated with our results with 60.46% substitutions are transitions with 57.69% between pyrimidine bases and 23.07% between purine bases. They concluded that this is similar to the mutation model of DNA oxidative decomposition caused by ROS in normal tissues. They concluded that the main cause of mtDNA mutations in tumors is the high level of ROS. It is noted a low content of Cytosine (C) in the third position of the codon. This shows that the transition from T to C is lower than that from C to T. Estimates of substitutions per site using the p- distance and distance to K2P have shown a greater rate of substitutions at the second codon position, differing from what is provided in the genetic code. Non-synonymous substitution rate which are higher than the rate of synonymous substitution per site. This is generally very rare. It has been observed that in human, dN>dS for immunoglobulin VH genes, MHC proteins and ribonucleases. The same phenomenon is observed for the virus proteins such as the envelope of the HIV virus or influenza virus hemagglutinin [23]. MTCYB is involved in the function of carcinogenesis by accumulating non- synonymous mutations that change the amino acid unlike synonymous mutations that alter codon but do not change the amino acid.
5. Conclusions
- Somatic mutations observed and which are specific to cancerous tissues and those common to cancerous tissue and control tissue with a significant difference could have a role in breast tumorigenesis. dN>dS, MTCYB is involved into the carcinogenesis by accumulating non-synonymous mutations. Our results indicate that alterations MTCYB are common in breast cancer.
ACKNOWLEDGEMENTS
- The authors are grateful to all Senegalese women who have voluntarily agreed to participate in this study as well as those who helped with the collection of samples.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML