-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Journal of Health Sciences
p-ISSN: 2166-5966 e-ISSN: 2166-5990
2012; 2(3): 19-22
doi: 10.5923/j.health.20120203.01
Antibiotic Resistance Profile of Gram Positive Bacteria Isolated from Wound Infections in Minna, Bida, Kontagora and Suleja Area of Niger State
Sani R. A.1, Garba S. A.2, Oyewole O. A.2, Ibrahim A.3
1Hospital Management Board, Minna, Niger State
2Department of Microbiology, Federal University of Technology Minna, Niger State
3College of Nursing Sciences, School of Midwifery, Minna, Niger State
Correspondence to: Oyewole O. A., Department of Microbiology, Federal University of Technology Minna, Niger State.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
Antibiotics resistance profiles of gram positive bacteria isolated from wound infections in four (4) General Hospitals (Bida, Kontagora, Minna and Suleja) in Niger State was carried out. Organisms isolated from surgical wounds were Staphylococcus aureus and Streptococcus pyogenes. Five hundred (500) samples (i.e. Two hundred (200) samples in Minna, One hundred (100) samples each from Suleja, Kontagora and Bida) of wound exudates from surgical wounds sites were analysed for their resistance pattern. From the five hundred (500) samples collected from all the locations, one hundred and twenty one samples (121) had Strept. pyogenes, one hundred and ninety seven (197) samples had S. aureus. S. aureus was more frequently isolated (62%) than Strept. pyogenes (38%) from wounds in all the locations. Both bacteria were tested for sensitivity to Tarivid, Pefloxacin, Ciprofloxacin, Augmentin, Gentamycin, Streptomycin, Ceporex, Nalidixic acid, Septrin, Ampicillin, ampiclox 30µg, zinacef 20µg, Amoxacillin, rocephin and erythromycin. Of the five hundred (500) wound samples from various locations 318 (64%) yielded growths while 182 samples (36%) yielded no growths. Most of all the isolates were sensitive to ciprofloxacin, pefloxacin and Tarivid while others were resistant to remaining antibiotics. S. aureus showed a higher resistance profile to most antibiotics used than Strept. pyogenes.
Keywords: Antibiotic, Resistance, Sensitive, Surgical Wounds, Gram Positive
Article Outline
1. Introduction
- All surgical wounds are contaminated by both pathogens and body commensals ranging from bacteria and fungi to other parasites[1-4]. The common gram positive organisms are the β – haemolytic streptococcus – Streptococcuspyogenes and Staphylococcus aureus. The gram negative aerobic rod is Pseudomonas aeruginosa. The facultative anaerobes include Enterobacter species, Escherichia coli, Klebsiella species and Proteus species. The fungi are Candida species and Aspergillus species[5,6], but the development of infection in the site depends on complex interplay of many factors[7]. These may be microbial virulence[1], patient risk factors like diabetes, cigarette smoking, obesity, and coincident remote site infections or colonization[8] and operation-related risk factors including prolonged hospital stay before surgery, duration of the operation, tissue trauma, poor homeostasis, and foreign materials in the wound. The presence of foreign materials increases the risk of serious infection even with relatively small bacterial inocu-lums[9]. The widespread uses of antibiotics, together with the length of time over which they have been available have led to major problems of resistant organisms, contributing to morbidity and mortality[4,10]. Antimicrobial resistance can increase complications and costs associated with procedures and treatment. Antimicrobial resistance among pathogens of wound infections is on the increase. The control of wound infections has become more challenging due to widespread bacterial resistance to antibiotics and to a greater incidence of infections caused by methicillin-resistant S. aureus (MRSA) and polymicrobic flora [4,10,11].Knowledge of the causative agents of wound infection in a specific geographic region will therefore be useful in the selection of antimicrobials for empiric therapy. The objective of the present study is to determine the antibiotic resistance profile of gram positive bacteria isolated from surgical wounds in Minna, Bida, Kontagora and Suleja areas of Niger State.
2. Materials and Methods
2.1. Collection of Samples
- Wounds samples were collected from five hundred (500) patients that undergo surgical operation in four (4) general hospitals in Minna, Bida, Kontagora and Suleja areas of Niger State. 200 samples were collected from general hospital in Minna while 100 samples were collected each from Bida, Kontagora and Suleja general hospitals. The wound types included boils, whitlow, abscesses, cervicitis, trauma wounds, burns, systemic ulcers, insect bites and swelling of no specific etiology. These samples were transferred to the Microbiology laboratory of Federal University of Technology, Minna for further analysis.
2.2. Characterization and Identification of the Isolates
- The collected samples were streaked on freshly prepared nutrient agar plates and incubated aerobically and anaerobically at 37℃ for 24 hours. Bacterial colonies differing in size, shape and colour were selected from the different plates and further subcultured on nutrient agar by the streak plate technique and incubated at 37℃ for 24 hours after which, were maintained on agar slants for further characterization and identification. The bacterial isolates were characterized based on colonial and cell morphology, growth on differential/selective media and biochemical tests which include Gram’s reaction, indole tests, methyl red, Voges-Proskauer, Citrate Utilization, Motility, endospore, utilization of carbohydrates such as glucose, sucrose, mannitol, lactose and fructose, oxidase, catalase, coagulase and starch hydrolysis test[12]. The bacterial isolates were identified by comparing their characteristics with those of known taxonomy using the schemes of[13].
2.3. Susceptibility of Isolates to Various Antibiotics
- Antibiotic sensitivity test were carried out on all isolates using paper (New Man England) disc diffusion technique. A total of 10 antibiotics were tested and 0.2ml of 12h peptone water culture of test organism was used to inoculate each organism on a dry sterile nutrient agar plate. The resistant profiles of bacteria isolated from surgical wounds were determined by standard methods. The antibiotic discs used are gram positive sensitive.
3. Results
3.1. Microorganisms Isolated from Samples at Each Location
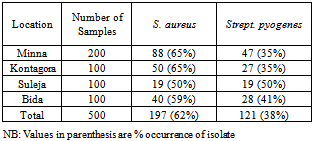
- Table 1 shows the gram positive bacteria isolated from wound samples in various general hospitals examined. S. aureus and Strept. pyogenes were the bacteria isolated. S. aureus had the higher occurrence in all four locations (62%) while Strept. pyogenes had occurrence of 38%.
3.2. Antibiotic Resistance of Bacteria in Minna
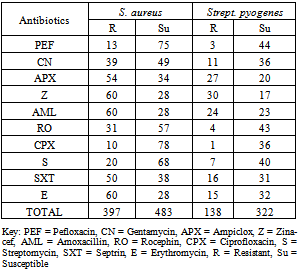
- Antibiotics resistance of bacteria isolated in Minna General Hospital is presented in Table 2. S. aureus had a total resistance of 397 while Strept. pyogenes had 138. S. aureus had a total susceptibility of 483 while Strept. pyogenes had 322 in all the antibiotics examined.
3.3. Antibiotic Resistance of Bacteria in Bida
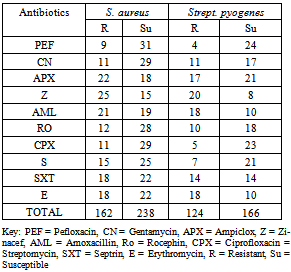
- Antibiotics resistance of bacteria isolated in Bida General Hospital is presented in Table 3. S. aureus had a total resistance of 162 while Strept. pyogenes had 124. S. aureus had a total susceptibility of 238 while Strept. pyogenes had 166 in all the antibiotics examined.
3.4. Antibiotic Resistance of Bacteria in Kontagora
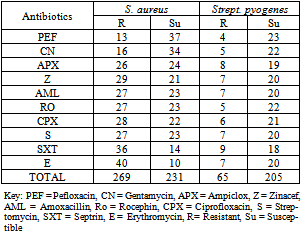
- Antibiotics resistance of bacteria isolated in Kontagora General Hospital is presented in Table 4. S. aureus had a total resistance of 269 while Strept. pyogenes had 65. S. aureus had a total susceptibility of 231 while Strept. pyogenes had 205 in all the antibiotics examined.
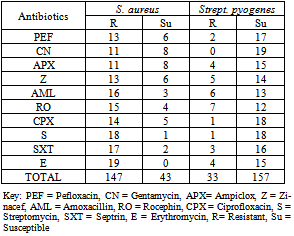
3.5. Antibiotic Resistance of Bacteria in Suleja
- Antibiotics resistance of bacteria isolated in Suleja General Hospital is presented in Table 4. S. aureus had a total resistance of 147 while Strept. pyogenes had 33. S. aureus had a total susceptibility of 43 while Strept. pyogenes had 157 in all the antibiotics examined.
|
4. Discussion
- Staphylococcus aureus is the leading cause of wound infection both surgical and accidental followed by S. epidemidis and they are pyogenic, meaning that they characteristically cause purulent discharge, otherwise known as pus[14]. Treatment of staphylococcal infection has been problematic because of the development of resistance to different antimicrobial medications by production of either plasmid encoded beta-lactamase, modification of penicillin binding proteins[14]. S. aureus showed resistance to Erythromycin, Zinacef, Amoxacillin, Ampiclox and Septrin in all the locations except in Bida. The percentage resistant ranged from 60 – 68%[15]. The ability of staphylococci to persist in adverse environments and their extraordinary potential to develop antimicrobial resistance may contribute to resistance patterns in sites of isolation and locations[16]. In the year 2000, a new method of reducing the problem of resistance was developed, that is using the combination of two substances that act synergistically. Despite the uses of synergistic antibiotics e.g. ampiclox, S. aureus was able to develop resistance to this drug in all the locations (Bida, Minna, Suleja and Kontagora). Also, the resistance pattern of S. aureus and S. epidermidis in Minna, Bida, Kontagora and Suleja is not in agreement with the study of[17].
|
|
|
|
5. Conclusions
- The findings of this study suggest that bacterial resistance in surgical wound infections is becoming serious menace in all the study area.Staphylococcus aureus is still the most frequently involved pathogen, showing high resistance rates of bacteria isolated from surgical wounds.Tarivid, ciprofloxacin and Pefloxacin are the best therapeutic options to treat these infections because of the less resistant caused by these organisms.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-Text HTML
Full-Text HTML