-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Frontiers in Science
p-ISSN: 2166-6083 e-ISSN: 2166-6113
2022; 12(1): 9-12
doi:10.5923/j.fs.20221201.02
Received: Aug. 19, 2022; Accepted: Sep. 7, 2022; Published: Sep. 15, 2022

Optical and Acoustic Resonance Coupling in DNA Molecules Theoretical Approach
Vitaly Vodyanoy
Department of Anatomy, Physiology and Pharmacology, Auburn University College of Veterinary Medicine, Auburn, Alabama, USA
Correspondence to: Vitaly Vodyanoy, Department of Anatomy, Physiology and Pharmacology, Auburn University College of Veterinary Medicine, Auburn, Alabama, USA.
| Email: |  |
Copyright © 2022 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

New molecular recognition signatures based on the terahertz/acoustic coupling that is complimentary to present infrared (IR) and ultraviolet (UV) technologies are described. They can provide increased detection, protection, decontamination, diagnostic and therapeutic capabilities. The system can provide increased discrimination of biological in complex environments. It is remote, fast, requires no labels or reagents, and it is very specific. This work will provide a better understanding of the fundamental acoustic/optical properties of DNA. This will provide a theoretical model of acoustic/optical coupling in DNA.
Keywords: Internal vibrations, Nucleic acid, Detection, DNA interrogation
Cite this paper: Vitaly Vodyanoy, Optical and Acoustic Resonance Coupling in DNA Molecules Theoretical Approach, Frontiers in Science, Vol. 12 No. 1, 2022, pp. 9-12. doi: 10.5923/j.fs.20221201.02.
Article Outline
1. Introduction
- The submillimeter-wave frequency range (0.01-10 THz) has been demonstrated to be potentially useful for molecular recognition [1,2]. This range belongs to the frequency gap between microwave and infrared radiation, where generation, propagation, and detection involve very complex and expensive technology [3]. According to theoretical predictions, DNA has a spectrum of internal vibrations consisting of high-frequency (THz) optical branches paired with relatively low frequency acoustic oscillations [4]. In this work, we explore acoustic/optical coupling for nucleic acid identification and characterization. When applied, this technology will help with the fast detection and identification of DNA. Coupling of the acoustic and THz modes will provide a new signature regime. While a unique signature at the level of species DNA will afford rapid identification, a deeper understanding of the fundamental properties of DNA will allow building more realistic theoretical models of nucleic acids. Elucidation of the acoustic/optical properties of DNA can provide the basis for improvements in measures for providing biological protection to live species.Here, we will characterize optic and acoustic coupling in DNA molecules and molecular signatures and develop a theoretical model of acoustic/optical coupling in DNA. We will discuss DNA interrogation principles for diagnostics and therapy. We will elucidate the mechanisms of DNA excitation in live cells and spores by EM/acoustic vibrations for the detection of pathogenic microorganisms by DNA interrogation.
2. Rationale and Methods
- A dual resonance of the high-frequency electromagnetic waves and acoustic waves occurs over the same length of the DNA molecule under the following simplified conditions:Fem/ Fac=Vem/Vac, where Fem and Fac are the resonance frequencies of electromagnetic and acoustic oscillations, respectively, and Vem and Vac are the velocities of light and sound in a DNA molecule, respectively. Different from THz spectroscopy, because the velocity of light in the DNA molecule is ~100,000 times larger than the velocity of sound [4], the acoustic resonance frequencies are in the MHz-GHz range. MHz-GHz acoustic spectroscopy is very well developed and is not as expensive as THz spectroscopy. A simple theoretical model of DNA composed of discrete coupled disks that have only longitudinal degrees of freedom can be described by the following dispersion equation [5]:
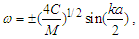 | (1) |
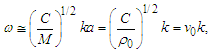 | (2) |
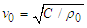 is the wave group velocity in the equivalent homogeneous string. The maximal value of k can be estimated as km=π/a. Therefore, the long wave limit of ω may be estimated from Eq. 2 as follows:
is the wave group velocity in the equivalent homogeneous string. The maximal value of k can be estimated as km=π/a. Therefore, the long wave limit of ω may be estimated from Eq. 2 as follows: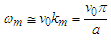 | (3) |
3. Optical and Acoustic Coupling
- Realistic DNA models predict the existence of very rich dispersion spectra consisting of a few optical and acoustic branches [4]. Since the frequency of phonons along the acoustic branches can become very small as the wavenumber decreases, resonance could occur at low frequency and high-frequency oscillations. The conditions for coupling EM fields to the phonon modes of double-stranded DNA through the electrical charges bound to the strands were carefully analyzed by Chitanvis and Golo [10,11]. They predicted that the application of EM fields near the natural phonon modes of DNA can create resonant amplification. It was shown by numerical simulation that even in the presence of solvent, the amplitude can grow by a factor of 50 after approximately 900 vibrational time periods if = the parameter values for irradiated frequency, electric field and coupling with solvent are met [11]. This means that pumping the external radiation causes the initial small amplitude to increase by many orders of magnitude. These publications [10,11] and earlier theoretical papers [12,13] were inspired by an important experimental work of Edwards et al., [14] who demonstrated the resonant adsorption of microwave energy by aqueous solutions of DNA of known lengths. Assuming adsorption peaks arise from resonances of longitudinal acoustic waves, the resonant frequency fn for a DNA molecule of length L (determined by the number of base pairs) is described by
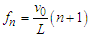 | (4) |
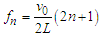 | (5) |
4. Proposed Experiment
- Here, we can use linear and circular DNA of various lengths, including full bacterial genomes and natural plasmids of individual bacterial species. For example, E. coli K-12 contains the plasmid pUC18. The plasmid pUC18 is responsible for the resistance of bacteria to the antibiotic ampicillin. Denature of the plasmid by induction of resonance energy can be easily verified by examining the receptivity of bacteria to ampicillin. When bacteria lose the plasmid, they cannot resist the antibiotic. If resistance to ampicillin of irradiated bacteria is reversed by conjugation with intact plasmids, then the interaction of EM oscillations with the plasmid of bacteria is specific. Both E. coli K-12 and plasmid pUC18 are commercially available. Signature spectra of DNA and bacteria will be analyzed, interpreted, and used for the theoretical model.DNA samples can be irradiated by THz sources in the range of 0.01-10 THz, and resonant absorption at fundamental and harmonic frequencies for both circular and linear nucleic acids will be measured by the detector. Additionally, one can measure the acoustic signals in DNA molecules using acousto-optic coupling similar to that utilized in the all-fiber acousto-optic tunable filter [15]. A liquid optical fiber filled with a solution of DNA can be subjected to THz EM and acoustic interrogation (Figure 1).
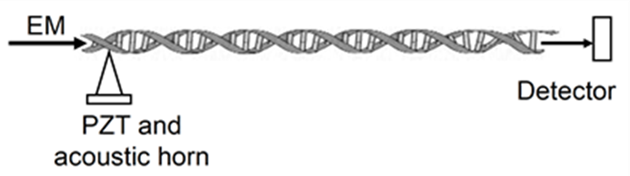 | Figure 1. Acousto-optic coupling |
5. Conclusions
- This system achieves results rapidly in real time and is label-free, single base sensitive, and inexpensive. This approach will help with the identification and characterization of DNA molecules; the interrogation of DNA by acoustic or optic oscillations; the diagnosis of defects in DNA molecules; the eradication of DNA of pathogenic bacteria; the correction of defective DNA; and the remote detection of pathogenic microorganisms in humans, animals, air, water, soil, and food products.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML