-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Frontiers in Science
p-ISSN: 2166-6083 e-ISSN: 2166-6113
2018; 8(1): 18-25
doi:10.5923/j.fs.20180801.03

Prevalence of Antibiotic Resistant and Biofilm Producing Escherichia coli and Salmonella spp from Two Sources of Water in Mubi, Nigeria
Musa Yakubu Tula, Grace Amara Onyeje, Ayuba John
Department of Biological Science Technology, Federal Polytechnic Mubi, Adamawa State, Nigeria
Correspondence to: Musa Yakubu Tula, Department of Biological Science Technology, Federal Polytechnic Mubi, Adamawa State, Nigeria.
| Email: |  |
Copyright © 2018 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

The presence of bacteria indicative of faecal pollution or potential pathogen in water meant for drinking or other domestic activities is of public health significance. The problem is further complicated when these organisms exhibit multiple antibiotic resistant phenotype in addition to other virulent factors like biofilm. A total of 72 water samples were analysed for the presence of Escherichia coli and Salmonella spp by standard bacteriological methods, antibiotic susceptibility, curing and biofilm production. These include 30 from 10 brands of sachet water and 42 from 21 boreholes in seven different wards of Mubi. Thirty (41.7%) E. coli and 13 (18.1%) Salmonella spp were isolated in all the 72 water samples analysed. From these, 2 (6.7%) and 28 (66.7%) E. coli were isolated from sachet and borehole water samples respectively. All Salmonella spp were isolated from borehole water samples. Both Escherichia coli and Salmonella spp exhibit high and variable antimicrobial resistance to most of the tested antimicrobials especially amoxicillin-clavulanic acid (AMC), cotrimoxazole (SXT) and chloramphenicol (CHL). Resistant to all the antibiotics was not statistically different (P=0.172) between E. coli and Salmonella spp. Resistant markers in both E. coli and Salmonella spp were cured variably. Biofilm was produced by 76.7% and 53.8% E. coli and Salmonella spp respectively. There was significant correlation in antibiotic resistance between biofilm and non-biofilm producing strains of E. coli (P=0.044) and Salmonella spp (P=0.015). The results indicate the presence of bacteria spp of public health significance in two sources of water in Mubi which possibly exhibit plasmid mediated resistance in addition to biofilm production.
Keywords: Prevalence, Salmonella, E. coli, Resistant, Biofilm, Water, Mubi
Cite this paper: Musa Yakubu Tula, Grace Amara Onyeje, Ayuba John, Prevalence of Antibiotic Resistant and Biofilm Producing Escherichia coli and Salmonella spp from Two Sources of Water in Mubi, Nigeria, Frontiers in Science, Vol. 8 No. 1, 2018, pp. 18-25. doi: 10.5923/j.fs.20180801.03.
Article Outline
1. Introduction
- Population growth and failure of government to provide portable pipe-borne water to communities led to over dependence on alternative sources of water especially bore hole, well and sachet water, some of which possessed questionable microbiological quality. Several studies have also shown that water harbor divers types of bacterial species which includes; E. coli, Salmonella Typhi, Bacillus, Staphylococcus, Enterobacter, Klebsiella, Pseudomonas spp, etc. [1, 2]. These organisms are of clinical and public health significance more especially Salmonella spp and E. coli.Most of the bacterial isolates found in water are opportunistic pathogens. Their presence in water meant for domestic activities call for concern because these microorganisms can lead to infection of certain segments of the population majorly the newborn babies, children and immunocompromised individuals [2].Salmonella infections may be divided into five categories: gastroenteritis, enteric fever, bacteraemia, localized infections, and the chronic carrier state. Salmonella disease has been linked to contaminated food and water supplies [3]. Escherichia coli is a member of the faecal coliform group and is a more specific indicator of faecal pollution than other faecal organisms [4]. Escherichia coli is a bacterium that is commonly found in the lower intestine of warm-blooded animals. Most strains are harmless. However, some strains are pathogenic; meaning that they have the potential to cause infections such as urinary tract infection, bacteraemia, pneumonia, bacterial meningitis, septic arthritis, severe bloody diarrhoea, which may in some cases result in an acute kidney failure requiring intensive care.The presence of Multi drug resistant Salmonella spp and Escherichia coli more especially from sources of water for drinking has been reported [5, 6]. The presence of antibiotics resistant bacteria in sources of water is worrisome because of the danger in promoting multiple antibiotic resistant organisms in humans. The problem is further compounded when these organisms possessed certain virulent factors such as biofilm production.Slime or Biofilm production could play an important role in the adherence of these microorganisms to mucous epithelia and this could be quite important for colonization, and hence, its virulence factor [7]. Biofilm forming strains adhere better to surfaces and have lower antibiotic susceptibility. Moreover, these traits make them better at eluding host defense mechanisms [8]. The ability of an organism to produce slime is significantly associated with its capability to produce recalcitrant illnesses [7] and that kind of organism is very difficult to eradicate [9].Mubi is an ancient town and the second largest city in Adamawa State, North-eastern Nigeria. However, the supply of pipe-borne water in Mubi is rather elusive for more than two decades. Hence, people in Mubi largely depend on other sources of water for drinking and other domestic purposes. One of such water source is borehole which serves as the major source of drinking water in Mubi and environs. Borehole water in Mubi metropolis is virtually vended in 20-25l gallon carried on hand push truck or open vans and water tankers. However, the major concern is that most of these boreholes were cited in unhygienic areas, closer to refuse dump sites.Another source of drinking water in Mubi is sachet water popularly known as ‘pure water’. The production, marketing and consumption of sachet water in Mubi have increased tremendously over the years with several brands being marketed. The perception is that water in sachet is pure and devoid of any contaminating indices. However, several reports showed that majority of water in sachet were of poor microbiological quality [10-12].The guideline set by the World Health Organization, states that quality drinking water must not contain Escherichia coli and other opportunistic pathogens [13]. The guideline further stated that water should be tested against the presence of highly virulent pathogens such as Salmonella spp, Shigella dysenteriae and Vibrio cholerae that are responsible for typhoid and gastroenteritis, bacillary dysentery and cholera diseases respectively which arises due to high level of organic decay and fermentation on tropical waters.Since water is universally consumed in large quantity and the potential of drinking water to transport microbial pathogens to great number of people causing subsequent illness is well documented in countries at all levels of economic development [14]. It is important to know and monitor the prevalence of bacteria of public health significance taken in by drinking water. Therefore, this study intends to elucidate the prevalence of E. coli and Salmonella spp in two sources of water in Mubi, their antibiogram and the correlation of biofilm production with antibiotic resistance among the tested isolates.
2. Materials and Methods
2.1. Study Area
- Mubi metropolis is a geo-political area comprising of two local government areas; Mubi North and Mubi South. The metropolis is located between latitudes 10° 05' and 10° 30'N of the equator and between longitude 13° 12' and 13° 19'E of the Greenwich meridian. The two Local government areas occupy a land area of 192,307 Km2 and support a total population 260,009 people (National Population Census 2006). The area shares international boundary with Cameroon Republic to the east and local boundary with Maiha L.G.A in the South, Hong L.G.A in the West, Michika L.G.A in the East. The major ethnic groups in Mubi includes; Gude, Fali, Kilba, Higgi, Marghi and Nzanyi [15].
2.2. Sample Collection
- A total of 72 water samples were collected and analyzed in this study. Out of these, 30 were sachet water belonging to 10 brands (replicated three times) while 42 water samples were collected from 21 different boreholes located in seven wards in and around Mubi. These wards include Nassarawo, Dirbishi, Vimtim, Monuva, Kolere, Wuro-patuji and Sabonlayi. In each ward, three boreholes were chosen and their samples replicated two times.The sachet water were coded as follows; OFR, HCM, SDK, KGN, AHS, DYJ, OYM, UDS, RNB and YNS. Samples were labeled and transported in ice packs to the laboratory and were analyzed within 1-2hrs after collection.
2.3. Isolation of Salmonella spp
- Six milliliters (6ml) from each sample was introduced into 15ml pre-enrichment broth (phosphate peptone water) and was incubated for 18-24hrs at 35-37°C. After incubation, 1.5ml of pre-enrichment culture was transferred aseptically into 10ml enrichment broth (tetrathionate broth). These were incubated aerobically for 24hrs at 37°C. A loopful of culture from each broth was streaked unto selective enrichment agar (Salmonella-Shigella agar and Deoxycholate citrate agar) and was incubated for 18-24hrs at 37°C. Suspected colonies (pale with dark centers) were further subjected to standard biochemical tests [16].
2.4. Isolation of Escherichia coli
- Five milliliters (5ml) of each water sample were introduced into test tubes containing sterilized Lactose and MacConkey broth. After incubation for 24hrs, a loopful from each tube was streaked into MacConkey agar (MCA) and Eosin methylene blue (EMB) agar. Isolates with green metallic sheen with dark centers on EMB were presumptively taken as E. coli, while lactose fermenters on MCA were further subjected to standard biochemical test.
2.5. Prevalence
- The prevalence of both E. coli and Salmonella spp. in water samples was calculated as (No. of samples from which pathogens were detected/total no. samples tested) × 100
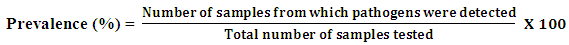
2.6. Antibiotic Resistance Profile
- Antibiotic susceptibility profiles of the bacterial isolates were evaluated using disk diffusion assay. The antibiotic discs containing the following antibiotics were used: cotrimoxazole (30μg), chloramphenicol (30 μg), sparfloxacin (10 μg), ciprofloxacin (10 μg), amoxicillin (30 μg), amoxicillin-clavulanic acid (30 μg), gentamycin (10 μg), perfloxacin (30 μg), ofloxacin (10 μg) and streptomycin (30 μg). The discs were aseptically placed on the surface of Mueller-Hinton agar (MHA) plates that has already being seeded with 0.5 McFarland standard of the test isolates and were incubated at 37°C for 18-24hrs. After incubation, diameters of zone of inhibitions were observed and measured in millimeters accordingly. The interpretation of the measurement as sensitive and resistant was made according to the manufacturer’s standard zone size interpretative table [17].
2.7. Curing Analysis
- Curing was carried out to determine the location (plasmid-borne or chromosomal) of the drug resistance marker(s). Elimination of the resistant plasmid was done using 10% acridine orange. The isolates were grown for 24hrs at 37°C in nutrient broth containing 10% acridine orange. After incubation, the broth was agitated to homogenize the content and loopful of the broth medium were then sub-cultured onto Mueller-Hinton Agar (MHA) plates and antibiotic sensitivity testing was carried out by disk diffusion method. Cured markers were determined by comparison between the pre- and post-curing antibiogram of each isolate [17].
2.8. Biofilm Formation
- The tube method was use to determine biofilm formation among the E. coli and Salmonella spp. Freshly grown colonies of the test isolates were introduced into 10 ml Trypticase Soy Broth. After incubation for 24hrs at 37°C, the tubes were washed with phosphate buffer saline, dried and stained with crystal violet (0.1%) for 20 min. The excess stain was washed with deionised water. Tubes were dried in an inverted position at room temperature. Biofilm formation was considered positive when a visible film lined the bottom of the tube. The isolates were grouped as non-biofilm producers (no visible film line), moderate-biofilm producers (medium intense film line) and high-biofilm producers (intense film line) [18].
2.9. Statistical Analyses
- Two-way analysis of variance (using Microsoft Excel) and correlation studies (using SPSS) were used to determine the contribution of biofilm to antibiotic resistance of the test isolates. Mann-Whitney statistics was used to determine differences in resistant phenotypes between E. coli and Salmonella spp. Significance difference was defined when p≤ 0.05.
3. Results
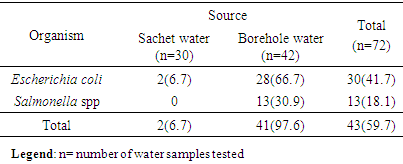
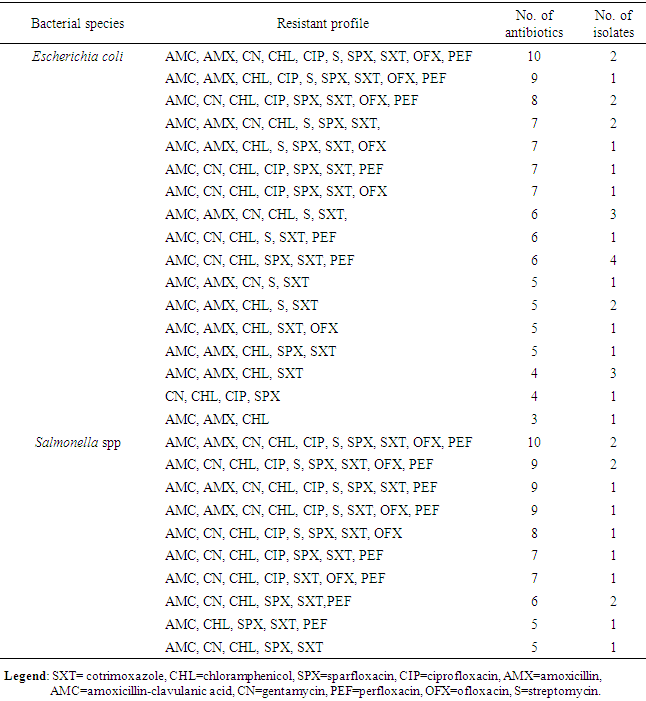
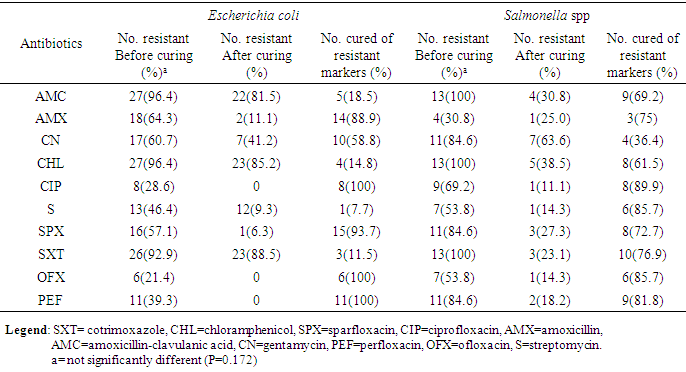
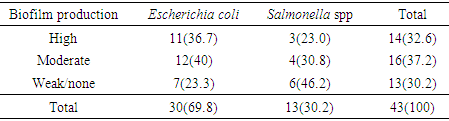
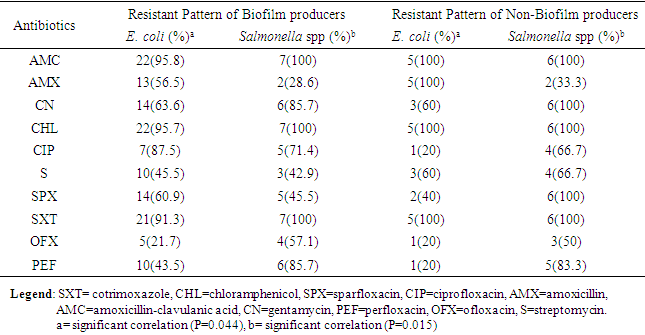
- The result in Table 1 showed the distribution of E. coli and Salmonella spp isolated from sachet and borehole water samples. Thirty (41.7%) E. coli and 13 (18.1%) Salmonella spp were isolated in all the 72 water samples analyzed. From these, 2 (6.7%) and 28 (66.7) E. coli were isolated from sachet and borehole water samples respectively. All Salmonella spp were isolated from borehole water samples. The result of the resistant pattern showed that E. coli were mostly resistant to amoxicillin-clavulanic acid (AMC; 96.4%), cotrimoxazole (SXT; 92.9%) and chloramphenicol (CHL; 96.4%) but least to ofloxacin (OFX; 21.4%) and ciprofloxacin (CIP; 28.6%). Moreover, E. coli exhibit 17 resistant profiles with AmcCnChlSpxSxtPef as the most resistant profile. In the same vein, Salmonella spp were 100% resistant to amoxicillin-clavulanic acid (AMC), chloramphenicol (CHL) and cotrimoxazole (SXT). Resistant to other antibiotics were also high and variable but least to amoxicillin (AMX; 30.8%) (Table 3). Salmonella spp exhibits 10 resistant profiles (Table 2). Resistant to all the antibiotics was not statistically different (P=0.172) between E. coli and Salmonella spp. Resistant markers in both E. coli and Salmonella spp were cured variably (Table 3). In E. coli, 100% were cured of their ciprofloxacin (cipr), ofloxacin (oflr) and pefloxacin (pefr) resistant markers. Moreover, 93.7% and 88.9% of E. coli were cured of their Sparfloxacin (spxr) and amoxicillin (amxr) resistant markers respectively. For Salmonella spp, 89.9%, 85.7%, 81.8% respectively were cured of their ciprofloxacin (cipr), Streptomycin (sr), Ofloxacin (oflr) and pefloxacin (pefr) resistant markers. Biofilm was produced by 76.7% and 53.8% E. coli and Salmonella spp respectively. From these, 36.7% and 23.0% were classified as high biofilm producing E. coli and Salmonella spp respectively. Also, 40.0% and 30.8% E. coli and Salmonella spp were moderate biofilm producers respectively (Table 4, Fig. 1). There was significant correlation in antibiotic resistance between biofilm and non-biofilm producing strains of E. coli (P=0.044) and Salmonella spp (P=0.015) (Table 5).
|
|
|
|
 | Figure 1. Tube biofilm formation among E. coli and Salmonella spp |
|
4. Discussion
- E. coli was found to be the most contaminant of the water samples examined in this work at the rate of 41.7%. According to the recommended drinking water standards, water should contain no bacteria pathogenic to humans; there is no tolerable lower limit for pathogens in water intended for consumption, preparing food, drink or for personal hygiene [19]. The prevalence of E. coli in this study was however similar to other results obtained from South-eastern Nigeria [20], North-western Nigeria [21] and South-western Nigeria [10]. The isolation of E. coli from sources of drinking water as shown in our study reflects poor sanitary conditions in the locations that were sampled. More worrisome, was the isolation from sachet water which was supposed to have undergone adequate treatment. Also, the presence of E. coli in the water samples was an indicator of possible faecal contamination of the water sources and the likely presence of other harmful organisms. This could possibly be due to treatment failure or unhygienic conditions with regard to sachet water [2]. It may also be due to improper construction of the boreholes, animal waste, proximity to toilet facilities, sewage, refuse dump site and various human activities surrounding the water sources [22]. A previous survey of Mubi metropolis showed that most boreholes are situated in less than 10m to septic tank and refuse dump sites [23]. In this study, Salmonella spp was not isolated from sachet water. However, the bacterium was isolated from borehole water samples with the prevalence of 18.1%. A study in South-eastern Nigeria reported prevalence rate of 30% in reservoir water and 7.5% in borehole water [6]. Lower prevalence rate of Salmonella spp from other water sources have been reported from other investigators such as 1.3% [24] from Yaoundé, Cameroun, 2.1% and 1.4% [25] from Madhyapur. Other researchers have reported higher prevalence rate of Salmonella spp from water sources other than borehole. These include; 71% [26] and 76.6% [27] prevalence rates. The isolation of Salmonella spp from borehole water samples from our study area means that the direct consumption of such water without treatment may be very dangerous. Salmonella spp is considered as the most common cause of food-borne infection throughout the world [27]. Others are responsible for enteric fever and septicaemia. Although the prevalence was low but the consumption of contaminated water with this bacterium may lead to infection. This is possible because Salmonella can survive for long period of time in water and can survive many challenges such as ultraviolet (UV) radiation from sunlight, poor nutrients, changes in pH and temperature [28, 29]. Moreover, when water contaminated with the bacterium is consumed directly, the bacterium voided with water can quickly pass through the stomach and then, enter to the intestines, without triggering the mechanisms of digestion and thereby the bacterium escapes the natural host defense mechanism. In Africa, the economic and health burden of water-borne diseases involving Salmonella spp is largely unknown because of the lack of comprehensive surveillance studies and credible measures of the disease incidence. However, a previous study documented the significant economic and health impacts associated with 2008 waterborne disease outbreaks involving Salmonella Typhimurium in Alamosa [30]. According to them, the estimated economic impact of the outbreak totaled over $2.6 million with an unanticipated loss of trust in the public water system after the outbreak. The presence of Salmonella spp in sampled borehole waters reflect possible contamination from broken underground pipes, flood water, farming activities, defecation and animal manures [5, 6, 31].From the above report, it is obvious that the presence of E. coli in sachet water brands and boreholes and Salmonella spp in borehole waters sold in Mubi metropolis could pose a serious threat to public health.High level of Salmonella antimicrobial resistance and E. coli as shown in our study was said to be responsible for reduced effectiveness of these drugs which are often used in the treatment of infections involving these organisms [32]. This could be due to improper use of drugs in hospitals and environment especially in animal husbandry. This may be enhanced by non-restricted and non-regulatory measure on the use of antibiotics. The finding of our study was contrary to previous studies in South-western Nigeria which reported low level of resistance to antimicrobials by E. coli and other enteric [33]. Another study [34] also showed that E. coli isolated from sources of water was highly sensitive to Chloramphenicol, amoxicillin-clavulanic acid and cotrimoxazole which was contrary to our findings.The presence of antibiotics resistant bacteria in water samples meant for drinking and other domestic activities is of public health significance. This is because the water samples can promote the growth and dissemination of antibiotic resistant organisms in humans and possible transfer of resistance genes to opportunistic pathogens and normal flora which may further complicate and increase the burden of antibiotic resistant organisms. This issue is of particular concern in developing countries where the common use of untreated, low quality water as drinking water sources together with inappropriate use and self-prescribing of antibiotics increase the risk of drug resistance [35].Curing analysis was carried out to determine the mechanism of antibiotic resistance portrayed by both E. coli and Salmonella spp. Although curing provides only the preliminary evidence that genetic traits are of extra-chromosomal nature, but loss of growth on antibiotic containing plate’s shows that the multi-drug resistance genes may be plasmid borne [17]. The curing result was variable in all the isolates. A previous study showed that partial curing as seen in our study occurs if copies of the plasmid lying closer to the membranes are completely eliminated by chemical agents while those lying closer to the nucleus may escape the curing effect [36].This implies that the genes coding for antibiotic resistance portrayed by E. coli and Salmonella spp in our study may be located on plasmids and under appropriate conditions, can be transferred to other bacteria through conjugation or other modes of recombination. The finding of our study also showed that majority of E. coli and Salmonella spp isolated from water sources are biofilm producers. Organisms with the ability to produce biofilm are of significant public health concern. This is because they exhibit dramatically decreased susceptibility to antimicrobial agents which can be intrinsic or acquired. Several studies showed that Salmonella spp and E. coli demonstrated the ability to produce biofilm on several surfaces [37-39]. This therefore explained the persistence of these organisms in the body of water analyzed. Our observations stem from the fact that biofilm can be beneficial for bacteria within a host; its presence also help to protect bacteria from environmental factors such as ultra violet radiation or desiccation [40, 41]. Also, biofilms are an ideal environment for the exchange of antibiotic resistance genes [42]. In this study, the antibiotic resistance of biofilm producing E. coli and Salmonella spp was found to correlate significantly with that of biofilm non-producing E. coli and Salmonella spp. This implies that there was no significant difference in antibiotic resistance between biofilm producing E. coli and Salmonella spp and non-biofilm producing E. coli and Salmonella spp. This observation was collaborated by previous study which reported similar observations in their studies [43]. However, our findings were contrary to the reports of other investigators which revealed that resistant to antibiotics was significantly higher in biofilm producing strains than non-biofilm producers [39, 44].
5. Conclusions
- This study, therefore, confirms previous reports that most of the bacteria isolates from water sources are becoming resistant to important antibiotics used by clinicians. Therefore, the high resistance pattern of E. coli and Salmonella spp in this work could be as a result of their acquisition of antibiotic resistance plasmids.
Authors’ Contribution
- Author MYT conceived and designed the study, carried out statistical analysis and wrote the first draft of the manuscript. Authors GAO and AJ facilitated sample collections in the field and analyses in the laboratory. All authors read and approved the final version of the manuscript.
ACKNOWLEDGEMENTS
- This work was supported by the Tertiary Education Trust Fund (TETFUND) of Nigeria (2011-2014 merged). Authors are therefore grateful.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML