Ahmed R. Sofy1, Abd El-Monem M. A. Sharaf1, Ali G. Al Karim2, Ahmed A. Hmed1, Kareem M. Moharam1
1Botany and Microbiology Department, Faculty of Science, Al-Azhar University, 11884 Nasr City, Cairo, Egypt
2Central Public Health Laboratories, Ministry of Health and Population, Cairo, Egypt
Correspondence to: Ahmed R. Sofy, Botany and Microbiology Department, Faculty of Science, Al-Azhar University, 11884 Nasr City, Cairo, Egypt.
| Email: |  |
Copyright © 2017 Scientific & Academic Publishing. All Rights Reserved.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Abstract
This observational and descriptive study was conducted to investigate and assess the microbiological quality and safety of meat and dairy food products in respective to the prevalence of Gram-negative harmful bacteria. For masterly-achievement of this target, a total number of 300 food samples were collected which included a beef burger, sausage, luncheon, turkey cheese and white cheese samples. Standard methods were used to determine Enterobacteriaceae count, coliform count and detection of Escherichia coli, Salmonella typhi, and Pseudomonas aeruginosa. Not all the food products sampled were found within acceptable safety limits, where these organisms were detected in some food products sampled. Luncheon samples showed high levels of positive results for Enterobacteriaceae, coliform, E. coli, S. typhi and P. aeruginosa, theincidence percentages were 56.6%, 35%, 81.6%, 23.3% and 18.33%, respectively. While, beef burger samples showed the lowest levels of Gram-negative prevalence as well as, S. typhi was not detected. In respective to the dairy products, the turkey cheese samples showed the highest incidence percentages of positive samples were 65%, 70%, 80%, 11.6% and 35% for Enterobacteriaceae, coliform, E. coli, S. typhi and P. aeruginosa, respectively. While white cheese samples showed the lowest levels. The distribution of Gram-negative bacteria in the various food samples examined, were E. coli (184 isolates) with 20, 28, 49, 48 and 39 isolates in a beef burger, sausage, luncheon, turkey cheese and white cheese samples, respectively. As well as, P. aeruginosa (51 isolates) with one, 5, 11, 21 and 13 isolates in the previous products samples, respectively. While, S. typhi showed the lowest dominant percentages (30 isolates) with 7, 14, 7 and 2 isolates in sausage, luncheon, turkey cheese and white cheese samples, respectively. The total number of bacterial isolates was 265 isolates. The results of the study indicate that most of the meat and the dairy food samples examined did not meet the quality standards, will render these foods unfit for human consumption. Also, the examined samples were not satisfactory in the course of public health standard as some pathogenic bacteria were detected. A sufficient number of these organisms will cause infection and intoxication, therefore, posing potential risks to consumers. Finally, the relevant authorities should draw the attention towards the health education campaign on food safety. As well as, food handlers should receive training on safety principles of good hygiene practice. Strict regulations in safe production, safe processing, and consumer awareness is highly recommended. People participation is a must.
Keywords:
Meat and Dairy Foods, Safety and Quality, Foodborne Pathogens, Gram-Negative Bacteria, Egypt
Cite this paper: Ahmed R. Sofy, Abd El-Monem M. A. Sharaf, Ali G. Al Karim, Ahmed A. Hmed, Kareem M. Moharam, Prevalence of the Harmful Gram-Negative Bacteria in Ready-to-Eat Foods in Egypt, Food and Public Health, Vol. 7 No. 3, 2017, pp. 59-68. doi: 10.5923/j.fph.20170703.02.
1. Introduction
Everyone faces the risk of contracting a foodborne illness simply because everyone eats. The World Health Organization (WHO) defines ‘Food Safety’ as the assurance that, food will not cause harm to the consumer when prepared and/or eaten in accordance with its intended use. Furthermore ‘Food Hygiene’ is defined, as all the measures necessary to ensure the safety, soundness, and wholesomeness of food at all stages of its production or manufacture until its final consumption [1]. Food industries worldwide have to conform to microbial standards associated with the safety and the quality of their products. Safety aspects are of major importance, thus are determined clearly and unambiguously. Usually, strict limits up to no tolerance are implemented for pathogenic microorganisms that could cause severe health problems to consumers [2].Over the last 25 years, the global incidence of foodborne infections has markedly increased, with nearly a quarter of the population at a high risk of illness [3]. The World Health Organization [1] estimates that foodborne and waterborne diarrheal diseases together kill around 2.2 million people annually. According to the Center for Disease Control and Prevention (CDC), each year millions of illnesses throughout the world can be traced to foodborne pathogens. Recently, in the USA, the Foodborne Diseases Active Surveillance Network (FoodNet) conducts surveillance at 10 U.S. sites for all infections caused by selected pathogens transmitted commonly through food. A total of 19,531 infections, 4563 hospitalizations, and 68 deaths associated with foodborne diseases were reported in 2012 [4]. The foods most commonly incriminated include meat and dairy products. Salmonella Typhimurium, Escherichia coli O157:H7 and Shigella spp. are among the major foodborne pathogens affecting people worldwide [5].All foodborne pathogens are important. However, they can be more or less hazardous based on different situations. For example, some foodborne pathogens cause serious illness with a low infectious dose, such as Salmonella spp. and E. coli O157:H7 [6]. Interestingly, Gram-negative bacterial pathogens account for approximately 69% of the cases of bacterial food-borne disease [7]. Except for listeriosis, almost all food-borne bacterial infections can be attributed to the ingestion of viable, gram-negative, enteric pathogens [7].Foodborne pathogens also lead to an economic burden every year. According to Scharff [8] in the USA 77.7 billion dollars was lost annually to investigate foodborne illnesses associated with 31 foodborne pathogens and spoilage microorganisms which lead to a serious economic loss.Although foodborne illnesses are caused by a wide range of foods, meat and dairy products industry remains the biggest safety concern and a focal point in many aspects. So, the objective of current study was conducted to evaluate the prevalence of the Gram-negative bacteria from these products at the end of their shelf-life. To give highlighted data throw more light where food regulatory jurisdictions should focus future educational activities. As well as, to assess food safety and maintenance public health by avoidance of the risk associated with such foods and can reduce economic losses by the early detection of inadequate food products.
2. Material and Methods
2.1. Samples Collection
During November 2016 and April 2017, a total of 300 food samples (180 meat products samples and 120 dairy products samples) were randomly collected from 40 different markets and specialty food shops from Cairo and Giza governorates in Egypt. The meat products samples were included: (60 Beef burger, 60 sausage, and 60 Luncheon samples). The dairy products samples were included: (60 Turkey cheese and 60 White cheese samples). All samples were collected in sterile plastic bags and transferred in ice boxes, and the samples were named, indexed and then investigated microbiologically.
2.2. Samples Preparation
Twenty-five grams of each sample were mixed and homogenized in the stomacher and diluted with (225 ml) buffered peptone water or sterile saline solution to make the sufficient dilutions for the microbiological analysis. Ten-fold dilutions of homogenates samples were prepared and inoculated onto appropriate media [9].
2.3. Isolation of Microorganisms from Meat and Dairy Samples
2.3.1. Detection and Enumeration of Enterobacteriaceae
The prevalence of Enterobacteriaceae was determined by spreading 0.1 ml of each sufficient dilution (dilution usually 10-1-10-4) on the surface of Violet Red Bile Glucose Agar (Oxoid; CM 485) and incubated at 37°C for 24 hr round purple colonies, surrounded by a purple halo, were considered to be Enterobacteriaceae [9].
2.3.2. Detection and Enumeration of Coliform Group
The coliform group was determined by spreading 0.1 ml of each sufficient dilution using a solid medium method onto plates of violet red bile agar media. The inoculated plates were incubated for 24 hr at 35°C. Coliform group to be counted will produce purple colonies surrounded by purple halos [9].
2.3.3. Isolation and Enumeration of E. coli
Carried out by streaking 0.1 ml of each of sufficient dilution of each food sample onto plates of Eosin Methylene Blue (EMB) (Oxoid; CM0069) and incubated at 37°C for 24 hr. Typical colonies (greenish metallic with dark purple center) were picked up and transferred to nutrient agar slants and incubated at 37°C for 24 hr for further identification [9].
2.3.4. Isolation of Salmonella
Aseptically 25 g of each sample was mixed with 225 ml of sterile buffer peptone water and incubated at 35°C for 24 hr. One to ten ml mixture was transferred to selenite cysteine broth and incubated at 35°C for 72 hr.Selective platting: Salmonella and Shigella (SS) agar (Oxoid; CM0099) and xylose lysine deoxycholate agar (XLD), (Oxoid; CM0469) plates were inoculated with enriched cultures then incubated at 37°C for 24 hr. Suspected colonies were creamy with or without black centers on SS agar and red with or without black centers on (XLD) agar [9].
2.3.5. Isolation and Enumeration of Pseudomonas spp
Twenty-five g of the sample was homogenized in 225 ml peptone water, and then serial decimal dilutions were prepared. Amount of 0.1 ml of each dilution was spread on Pseudomonas cetrimide, nalidixic acid (CN) agar; Pseudomonas agar base contains 10 ml/l glycerol and selectivity made by inclusion of cetyl trimethyl ammonium bromide (cetrimide; 200 mg/l) and nalidixic acid sodium salt 15 mg/l and were incubated at 25°C for 48 hr. All colonies that developed on the medium were counted and confirmed their identity as Pseudomonas by microscopically and biochemical examinations [10].
2.4. Purification and Identification of Bacterial Isolates
Bacterial colonies obtained were purified by a streak-plate method on nutrient agar medium. Pure isolates were maintained on slants of the same medium at 4°C for subsequent identification.
2.4.1. Morphological and Biochemical Identification
Almost all microscopically examinations and biochemical testing used for identification were carried out according to Bergey'smanual [11], Collins and Lyne [12] and Cheesbrough [13].
2.4.2. Identification by Analytical Profile Index (API) Strips
The Analytical Profile Index (API) strips (API 20E) obtained from (bioMérieux, France), were used as a biochemical system for identification of Enterobacteriaceae and other Gram-negative rod bacteria. The API strip consists of micro-tubes containing dehydrated substrates. These tests are inoculated with a bacterial suspension that reconstitutes the media. The strips were incubated for 18-24 hr at 37°C. During incubation; metabolism produces color changes that are either spontaneous or revealed by the addition of reagents. The reactions were read according to the reading table, and the identification was obtained by referring to the Analytical Profile Index [14].
2.5. Antibiotic Sensitivity Test for Bacterial Isolates
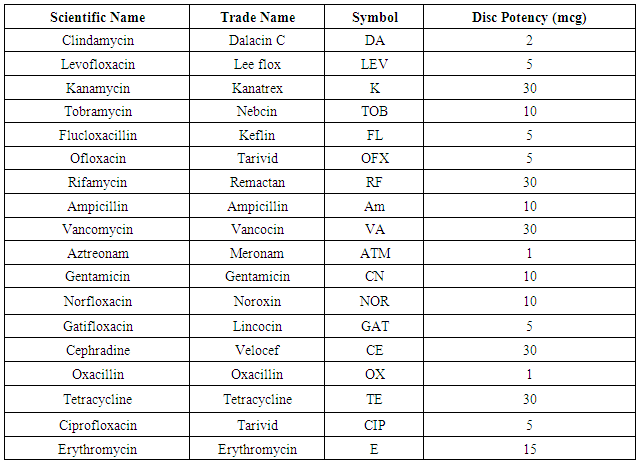
In this investigation, 18 commercially prepared antibiotic discs (6 mm in diameter) belonging to different groups were chosen for investigating their potency against bacterial isolates. The discs were obtained from (Oxoid, UK.) properties of the target antibiotics are listed in the table (1). In this test, the standard Kirby-Bauer disk diffusion method according to Bauer et al. [15] was performed in which, four to five similar colonies from overnight growth plates were transferred aseptically in sterile distilled water and vigorously agitated to give a turbidity that matches the 0.5 McFarland standard (approximately 108 cfu/ml) according to D’Amato and Hochstein [16].Table 1. Antibiotics for sensitivity test
 |
| |
|
Within 15min, sterile cotton swab dipped into the culture suspension was used for inoculating the surface of solidified Mueller-Hinton agar plates [17]. Antibiotic discs were dispensed onto the inoculated plate surface and gently pressed down using a sterile forceps to ensure complete contact with the agar. Within 15 minutes of applying discs, the inverted plates were aerobically incubated at 37°C for 24 hr. The resulted diameters of inhibition zones around the antibiotic discs were measured to nearest whole mm and interpreted according to protocols standardized for the assay of antibiotic compounds as guided by the National Committee for Clinical Laboratory Standards “NCCLS.” The results were categorized as R (resistant), I (intermediate sensitive), and S (sensitive) [17, 18].
3. Results and Discussion
3.1. Incidence of Gram-Negative Bacteria in Meat Products
The obtained results of the incidence of Gram-negative foodborne bacteria were summarized in table (2) and illustrated in figure (1). The mean value of Enterobacteriaceae; count on a beef burger, sausage and luncheon samples were 3.21, 3.77 and 4.55 cfug−1, respectively, with incidence percentages of positive samples were 30, 38.3 and 56.6%, respectively. The average count of the total Coliform group; on a beef burger, oriental sausage and luncheon samples were 3.36, 4.61 and 4.89 cfug−1, respectively, contained with contamination percentages 20, 28.3 and 35%, respectively (Table 2 & Figure 1). These findings corroborate previous works [19] found the hamburgers were categorized as unsuitable for human consumption in 31.4% of samples, with those testing positive for Enterobacteriaceae and coliform. The presence of Enterobacteriaceae coupled with unhygienic surroundings like sewage, improper waste disposal system, might be the possible sources of food contamination. Members of the family Enterobacteriaceae have been considered a potent cause of foodborne outbreaks, therefore; the presence of members of Enterobacteriaceae in the foods under study might pose a health risk to children and individuals with underlying conditions [20].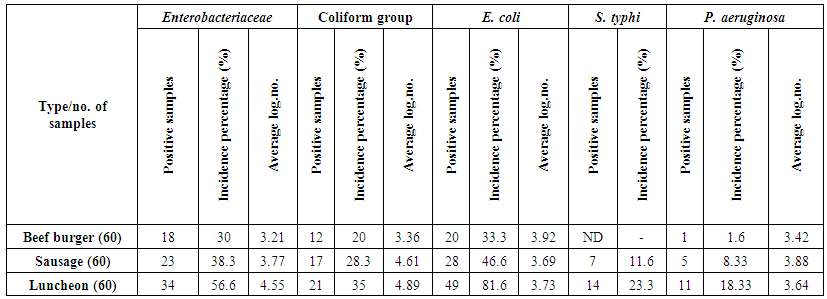 | Table 2. Incidence of Gram negative bacteria in different meat products |
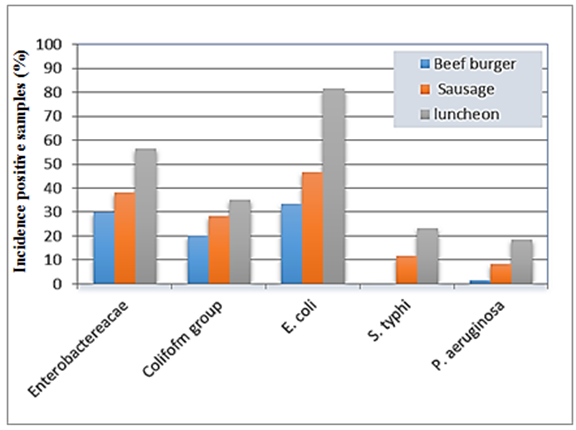 | Figure 1. Histogram of the prevalence of isolated Gram-negative foodborne bacteria from meat products |
E. coli count; the average count of E. coli was 3.92, 3.69 and 3.73 cfug−1 with incidence contamination percentages of positive samples 20, 28 and 49% of beef burger, sausage and luncheon samples, respectively (Table 2 & Figure 1). The prevalence of E. coli could be attributed to the use of contaminated water during the different stages of processing, in this respect the initial contaminated water used for washing the raw meat is also used for washing hands and utensils used in production, water is a major means by which E. coli are spread [21].Salmonella typhi was found in low-level contamination of meat products. Where, contamination percentages ranged from 0 to 14%, according to the type of meat product, the number of positive samples was 7 and 14 samples with incidence percentages 11.6% and 23.3% of sausage and luncheon samples, respectively, while the microorganism not detected in beef burger (Table 2 & Figure 1). Unwashed hands of an infected food handler may also contaminate food, and improper preparation and handling of foods at food service establishments are primary factors for Salmonella outbreaks [22]. These results are in harmony with Zhao et al. [23], they isolated in 19–54% of cattle carcasses, 1.9% of beef samples at retail and 4.2% of retail chicken samples. During weeks 1-52, 2002, there were 49 cases as of January 16, 2003, of salmonellosis in Trinidad and Tobago [24]. Lengeler et al. [25] reported the presence of Gram-negative facultative anaerobes including Klebsiella (K. penumoniae), Salmonella (S. typhi) and Proteus (P. vulgaris) in investigated food samples.Pseudomonas aeruginosa was detected in only one sample of beef burger, while oriental sausage and luncheon samples showed contamination with incidence percentages were 8.33% and 18.33%, respectively. The mean count of the microorganism in the different samples was 3.42, 3.88 and 3.64 cfug−1. The luncheon samples were exhibited the highest contamination percentage with E. coli, S. typhi and P. aeruginosa (Table 2 & Figure 1). Our findings are in an appositive relationship with the previous results obtained by Samson et al. [26], they reported, the presence of Gram-negative aerobes including Campylobacter (C. jejuni and C. coli), Pseudomonas (P. aeruginosa, P. fluorescens, and P. putida) in different meat samples examined.
3.2. Incidence of Gram-Negative Bacteria in Dairy Products
The results presented in table (3) and illustrated in figure (2) showed the incidence of Gram-negative foodborne bacteria in dairy samples. These results exhibited that, the average count of Enterobacteriaceae; were 4.46 and 3.82 cfug−1 with incidence percentages of positive samples 65 and 31.6% of turkey cheese and white cheese samples, respectively. 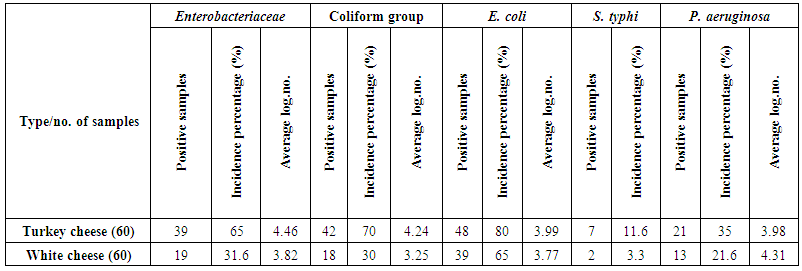 | Table 3. Incidence of some Gram-negative bacteria in dairy products |
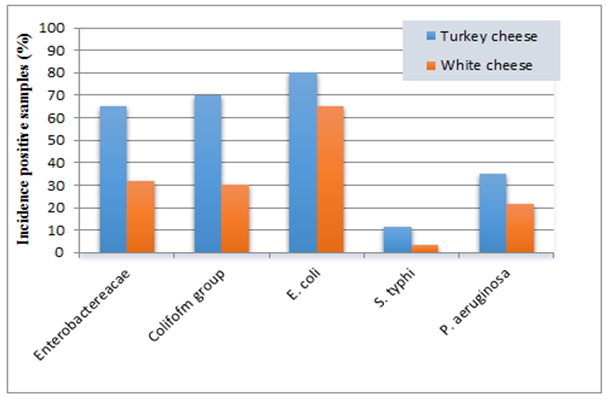 | Figure 2. Histogram of the prevalence of isolated Gram-negative bacteria from dairy products |
Coliform group count; the results showed that the average count was 4.24 and 3.25 cfug−1 for turkey cheese and white cheese samples, respectively. The highest contamination percentage was observed in turkey cheese (70%), while the incidence contamination percentage of white cheese samples was 30% (Table 3 & Figure 2). The incidence of Enterobacteriaceae and Coliform group is useful indicators of hygiene of post processing contamination of processed foods as those bacteria coming from equipment's or contact with raw foods. Similar results were observed by Hassan et al. [27], they showed that the mean value of Enterobacteriaceae count ranged from 3.9x102 to 1x103 for yogurt and Feta cheese. Also, the contamination percentage of the coliform group, are in consistent with Mohammed et al. [28], where showed that, the mean of coliform log no., was 6.28 cfug−1, while the contamination percentage with coliform was 73.33% of investigated samples. E. coli count; the mean count contained in turkey cheese and white cheese samples were 3.99 and 3.77 cfug−1 with incidence contamination percentages 80 and 65%, respectively (Table 3 & Figure 2). These findings corroborate with previous other studies in Egypt, where the pathogenic E. coli O157:H7 has been isolated from 19% of the total white cheese samples [29].S. typhi; showed contamination percentage 11.6% of turkey cheese samples and 3.3% of white cheese samples (Table 3 & Figure 2). The presence of Salmonella spp. in dairy products may be due to the using of raw milk for production accompanied by improper sanitary practices during manufacturing, handling and selling. The isolation of this pathogen hence these foods could be of high risk in transmitting enteric pathogens. These results are supported by the findings of De Buyser et al. [30], they found that Salmonella spp., were responsible for 29 outbreaks.P. aeruginosa; was detected in 21 samples of turkey cheese, with incidence contamination percentage 35% and mean count 3.98 cfug−1. While in white cheese, the number of contaminated samples were 13 samples with incidence percentage 21.6% and mean count 4.31 cfug−1 (Table 3 & Figure 2). These results are in consistence with the previous study by Leriche et al. [31], they found that, thirty Pseudomonas spp. strains isolated from milk, water, cheese center and cheese surface in two traditional workshops manufacturing raw milk.
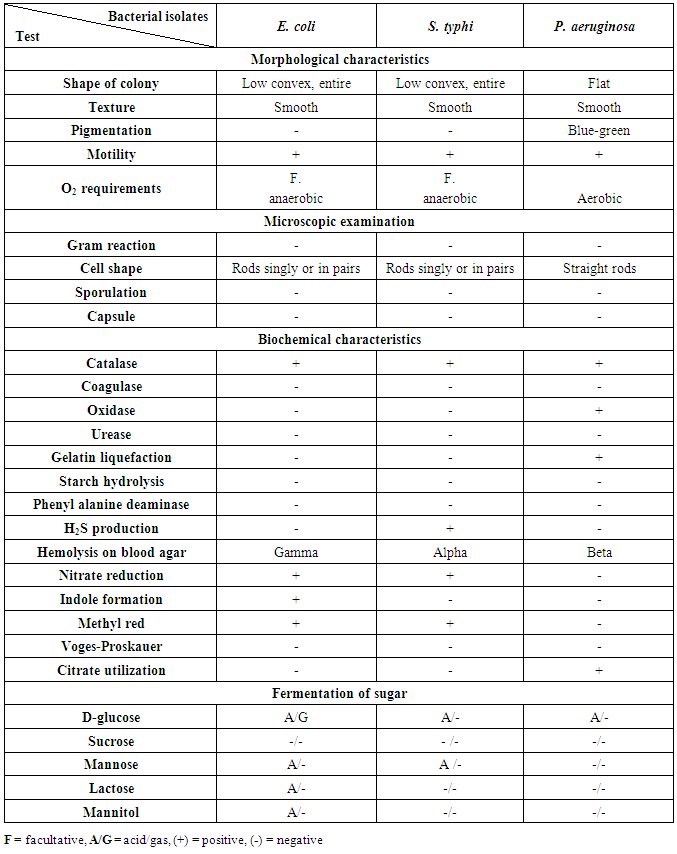
3.3. Identification of Bacterial Isolates
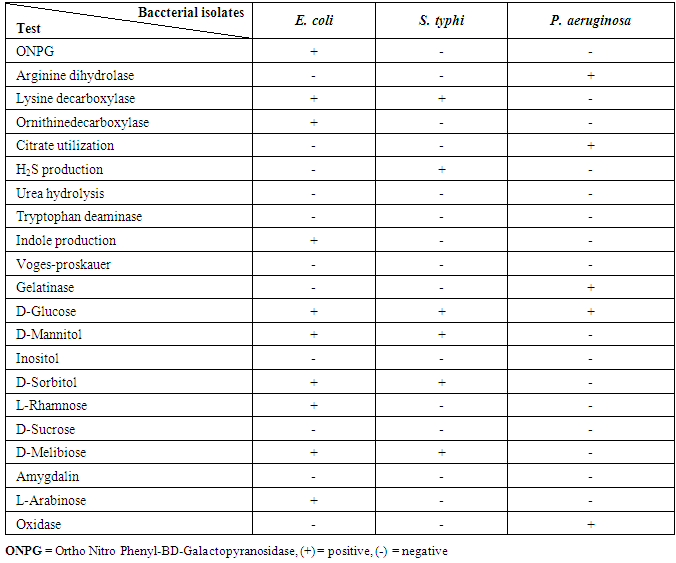
The bacterial isolates from meat and dairy food samples were taken to be identified according to their morphological, cultural characteristics and consumption of broth manual some biochemical tests according to Bergey's manual [11]. The results obtained from biochemical identification indicated that there are three different groups of foodborne bacterial isolates. The results of morphological and biochemical characteristics of bacterial isolates were given in table (4). The identified bacterial isolates from all collected food samples (meat & dairy) products belonged to two main bacterial families Enterobacteriaceae and Pseudomonadaceae. From each group, we take one isolate for confirming the identification of isolated pathogenic bacteria by using the Analytical Profile Index (API) system (Table 5).Table 4. Morphological and biochemical characteristics of bacterial isolates
 |
| |
|
Table 5. API identification of E. coli, S. typhi and P. aeruginosa
 |
| |
|
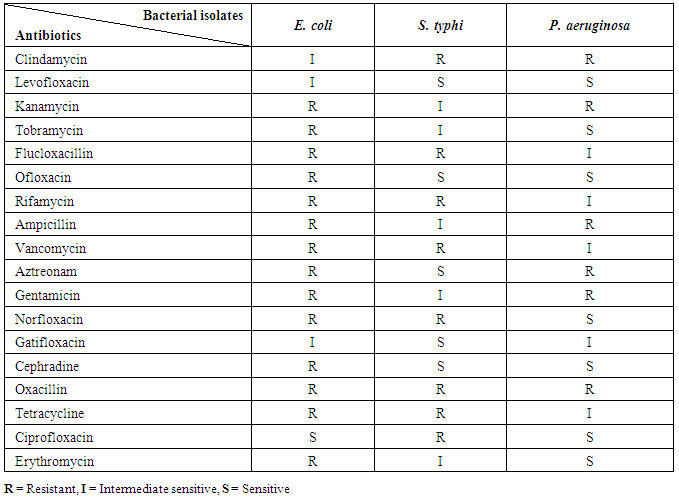
3.4. Antibiotic Sensitivity of Foodborne Bacterial Isolates
The antibiotic sensitivity of tested bacterial isolates showed different susceptibilities ranging from sensitive, intermediate and resistant against different tested antibiotics as indicated in the table (6).Table 6. Antibiotic sensitivity profiles of foodborne bacterial isolates
 |
| |
|
3.5. Prevalence of Pathogenic Foodborne Bacteria in Various Food Types
The results of morphological and biochemical characteristics of bacterial isolates from all collected food samples (meat & dairy) products belonged to two main bacterial families (Enterobacteriaceae and Pseudomonadaceae) depicts the occurrence of possible pathogens in the 300 food samples tested. Bacterial growth was observed in all the food types; the most prevalent bacteria were E. coli (184) isolates, P. aeruginosa were 51 isolates, while S. typhi exhibited the lowest dominant with (30) isolates (Figure 3). 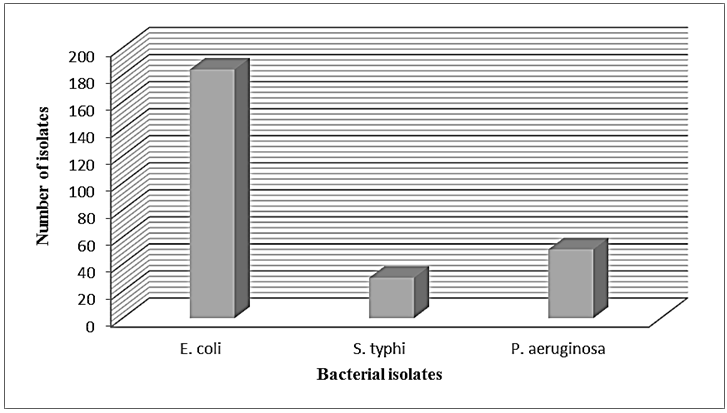 | Figure 3. Total number of isolated bacteria from meat and dairy products |
3.6. Distribution of Pathogenic Foodborne Bacteria in Various Food Types
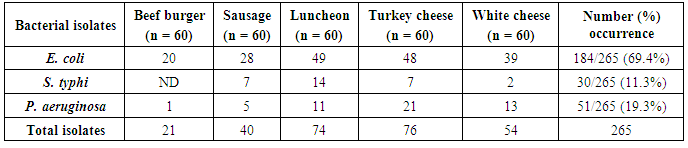
The results summarized in table (7) and illustrated in figure (4) showed the proportional distribution of foodborne bacterial isolates associated with meat and dairy samples examined. Since, E. coli was (184) with (20), (28), (49), (48) and (39) isolates in a beef burger, sausage, luncheon, turkey cheese and in white cheese samples, respectively.As well as, P. aeruginosa was (51) isolates with (1), (5), (11), (21) and (13) isolates in a beef burger, sausage, luncheon, turkey cheese and white cheese samples, respectively. While, S. typhi showed the lowest dominant percentage (30 isolates) with (7), (14), (7) and (2) isolates in oriental sausage, luncheon and turkey cheese and white cheese samples, respectively, while not detected in beef burger samples.These trends of results are harmony with Stopforth et al. [32], they Analyzed 1,022 fresh beef samples for levels of microbial populations (total aerobic plate count, total coliform count, and E. coli count) and the presence or absence of E. coli and Salmonella. The mean incidence rates of E. coli and Salmonella on raw beef cuts were 0.3 and 2.2%, respectively. Levels of the total coliform count and E. coli count did not (P > or = 0.05) appear to be associated with the presence of E. coli and Salmonella on fresh beef cuts. While our results were not accordance with Nyenje et al. [33], they assessed the microbiological quality of 252 samples which included rice, pies, beef and chicken stew, the organisms isolated included: Listeria spp. (22%), Enterobacter spp. (18%), Aeromonas hydrophila (12%), Klebsiella oxytoca (8%), Proteus mirabilis (6.3%), Staphylococcus aureus (3.2%) and Pseudomonas luteola (2.4%). Interestingly, Salmonella spp. and Escherichia coli were not isolated in any of the samples, the total number of bacterial isolates were 588 isolates.Table 7. Bacteria distribution in the various food samples examined
 |
| |
|
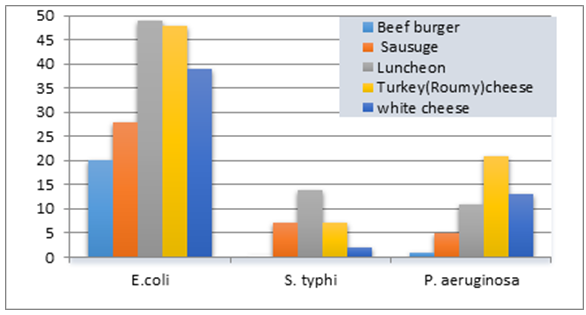 | Figure 4. Histogram of positive samples with foodborne bacteria of meat and dairy products |
References
| [1] | WHO (2010). Sixty-third World Health Assembly. Document WHA63/A63/11. Available at http://apps.who.int/gb/ebwha/pdf_files/WHA63/A63_11-en.pdf (Accessed 12th April 2013). |
| [2] | EFSA, European Food Safety Authority (2010). Trends and sources of zoonoses and zoonotic agents and food-borne outbreaks in the European Union in 2008. EFSA Journal 8(1): 1e368. |
| [3] | Oliver SP, Jayarao BM and Almeida RA (2005). Foodborne pathogens in milk and the dairy farm environment: food safety and public health implications. Foodborne Pathog. Dis., 2: 115–129. |
| [4] | CDC, (2013). Incidence and trends of infection with pathogens transmitted commonly through food — foodborne diseases active surveillance network, 10 U.S. sites, 1996–2012. Wkly Rep., 62(15): 283–287. |
| [5] | CDC (2010). Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 states, 2009. Morb. Mortal. Wkly Rep., 59: 418–422. |
| [6] | Kothary MH and Babu US (2001). Infective dose of foodborne pathogens in volunteers: a review. Journal of Food Safety, 21(1): 49-68. |
| [7] | Greig JD and Ravel A (2009). Analysis of foodborne outbreak data reported internationally for source attribution. Int. J. Food Microbiol., 130(2): 77–87. |
| [8] | Scharff RL (2012). Economic burden from health losses due to foodborne illness in the United States. J. Food Prot., 75(1): 123–131. |
| [9] | "FDA" Food and Drug Administration (2002). Bacteriological analytical manual. 9th Ed., AOAC Int., Arlington, VA, USA. |
| [10] | Roberts D and Greenwood M (2003). Practical food microbiology. 3rd edition. Blackwell Publishing Ltd, UK. 273-274. |
| [11] | Bergey’s Manual (2009). Bergey’s manual of systematic bacteriology. Sneath, P.H.A.; Mair, N.s.; Sharpe, M. Elizabeth and Holt, J.G. (Eds.) Pub. Williams and Wilkins, 2605. |
| [12] | Collins CH and Lyne PM (2004). Microbiological Methods, 8th ed. Arnold, London. |
| [13] | Cheesbrough M (2006). District laboratory practice in tropical countries, 2nd ed., Cambridge university press, New York. |
| [14] | Juang DF and Morgan JM (2001). The applicability of the API 20E and API rapid NET systems for the identification of bacteria from activated sludge. Elect. J. Biotech., 4(1): 1-7. |
| [15] | Bauer AW, Kirby WM, Sherris JC and Turck M (1966). Antibiotic susceptibility testing by a standardized single disk method. Tech. Bull. Regist. Med. Technol., 36(3): 49–52. |
| [16] | D'Amato RF and Hochstein L (1982). Evaluation of a rapid inoculum preparation method for agar disc diffusion susceptibility testing. J. Clin. Microbiol., 15(2): 282–285. |
| [17] | "NCCLS/CLSI" National Committee for Clinical Laboratory Standards/ Clinical and Laboratory Standards Institute (2007). Performance standards for antimicrobial susceptibility testing; Seventeenth informational supplement, M2-A9 and M7-A7. Wayne, P. A., U.S.A. |
| [18] | Hindler J (1998). Antimicrobial susceptibility testing. In: Isenberg, H.D. (Ed.). Essential Procedures for Clinical Microbiology. ASM press, Washington. |
| [19] | Bezerra ACD, Reis RB and Bastos DHM (2010). Microbiological quality of hamburgers sold in the streets of Cuiabá-MT, Brazil and vended hygiene-awareness. Ciênc. Tec. Aliment., 30(2): 121–132. |
| [20] | Centinkaya F, Cibik G, Soyuteniz E, Ozkin C, Kayali R and Levent B (2008). Shigella and Salmonella contamination in various foodstuffs in Turkey. J. Food Control, 19: 1059–1063. |
| [21] | Ologhobo AD, Omojola AB, Ofongo ST, Moiforay S and Jibir M (2010). Safety of street vended meat products-chicken and beef suya. African J. Biotec., 9(26): 4091–4095. |
| [22] | Jay JM (1992). Foodborne gasteroenteritis caused by Salmonella and shigella. Modern Food Microbiol. pp. 507–526. Chapman and hall, New York. |
| [23] | Zhao C, Ge B, De Villena J, Sudler R, Yeh E, Zhao S, White DG, Wagner D and Meng J (2001). Prevalence of Camplyobacter spp., Escherichia coli, and Salmonella serovars in retail chicken, turkey, pork and beeffrom the greater Washington, D.C. area. Appl Environ Microbiol, 67(12): 5431-5436. |
| [24] | CAREC (2003). Report on communicable disease. Caribb-ean Epidemiology Surveillance (CAREC) Report, 23(1): 12–18. |
| [25] | Lengeler JW, Drews G and Schlegel HG (2004). Biology of the prokaryotes, Blackwell Science, New York, pp. 80–110. |
| [26] | Samson RA, Hoekstra ES, Frisvad JC and Filtenborg O (2000). Introduction of food Microbio-logy and Airborne Fungi, 2nd ed., CBS Publications, The Netherlands. |
| [27] | Hassan NS, Esmail AS and Mahmoud AH (2009). Bacteriological assessment of some ready-to-eat foods. Kafr El-Sheikh, Vet. Med. J. Sci. Congress, 3: 474–487. |
| [28] | Mohammed HM, Ali M, Habib M, Servatkhah F and Mohammed RS (2001). Microbiological contamination of traditional chocolate ice cream sold in the Northwest region of Iran. Global Veterinaria, 6(3): 269–271. |
| [29] | Hosny IM, El Kholy WI, El Dairoty RK, El Shenawy MA and Sahar HS (2011). Microbiological quality of different varieties of ready-to-foods retailed in Cairo area. J. Am. Sci., 7(5): 527–536. |
| [30] | De Buyser ML, Dufour B, Maire M and Lafarge V (2001). Implication of milk and milk products in food-borne diseases in France and in different industrialized countries. Int. J. Food Microbiol., 20(1–2): 1–17. |
| [31] | Leriche F, Bordessoules A, Fayolle K, Karoui R, Laval K, Leblanc L and Dufour E (2004). Alteration of raw-milk cheese by Pseudomonas spp.: monitoring the sources of contamination using fluorescence spectroscopy and metabolic profiling. J. Microbiol. Methods, 59(1): 33-41. |
| [32] | Stopforth JD, Lopes M, Shultz JE, Miksch RR and Samadpour M (2006). Microbiological status of fresh beef cuts. J. Food Prot., 69(6): 1456-1459. |
| [33] | Nyenje ME, Odjadjare CE, Tanih NF, Green E and Ndip RN (2012). Foodborne pathogens recovered from ready-to-eat foods from roadside cafeterias and retail outlets in alice, eastern cape province, south africa: public health implications. International Journal of Environmental Research and Public Health, 29(8): 2608-2619. |









 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML



