-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Computer Science and Engineering
p-ISSN: 2163-1484 e-ISSN: 2163-1492
2016; 6(1): 14-18
doi:10.5923/j.computer.20160601.03

Fully Automated Adaptive Shape Segmentation Method for Noisy Complenentary DNA Microarray Images
Mai S. Mabrouk1, Samir Y. Marzouk2, Islam A. Fouad1
1Biomedical Engineering Department Misr University for Science and Technology (MUST), Egypt
2Basic and Applied Science, Faculty of Engineering, Arab Academy of Science and Technology, Al-Horria, Heliopolis, Cairo, Egypt
Correspondence to: Mai S. Mabrouk, Biomedical Engineering Department Misr University for Science and Technology (MUST), Egypt.
| Email: |  |
Copyright © 2016 Scientific & Academic Publishing. All Rights Reserved.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Due to the vast success of bioengineering techniques, a series of large scale analysis tools has been developed to discover the functional organization of cells. Among them, cDNA microarray has emerged as a powerful technology that enables biologists to study thousands of genes simultaneously within an entire organism, and thus obtain a better understanding of the gene interaction and regulation mechanisms involved. The analysis of DNA microarray image consists of several steps; gridding, segmentation, and quantification that can significantly deteriorate the quality of gene expression information, and hence decrease our confidence in any derived research results. Thus, microarray data processing steps become critical for performing optimal microarray data analysis and deriving meaningful biological information from microarray images. Segmentation is the process, by which each individual cell in the grid must be selected to determine the spot signal and to estimate the background hybridization. In this paper, a proposed segmentation method is explored; “Adaptive Shape Segmentation”. By inspecting the results, it was found that the proposed segmentation method can segment noisy microarray images correctly, gives high accuracy results and minimal processing time, and can be applied to various types of noisy microarray images.
Keywords: Noisy Microarray Image, Segmentation, Genes, Spot, Data Analysis, Adaptive Shape
Cite this paper: Mai S. Mabrouk, Samir Y. Marzouk, Islam A. Fouad, Fully Automated Adaptive Shape Segmentation Method for Noisy Complenentary DNA Microarray Images, Computer Science and Engineering, Vol. 6 No. 1, 2016, pp. 14-18. doi: 10.5923/j.computer.20160601.03.
Article Outline
1. Introduction
- Microarray technology came on time to cover the need to monitor in parallel all the DNA sequences and to have the adequate sensibility to detect the variation of gene expression. There may be tens of thousands of spots on an array. Each spot contains tens of millions of identical DNA molecules with lengths from tens to hundreds of nucleotides. Afterwards, the microarray slide is exposed to a set of labeled cDNA samples, which are derived from tissue of interest. With the completion of hybridization reaction, the amount of the target that bounds to each sample is measured with the aid of image capturing devices and computer technology. The measurement is based on the intensity of the spot. There are three basic steps in the processing of microarray image [1]; gridding, segmentation, and quantification. Image segmentation is the process of distinguishing objects from their background [2]. It is usually the first step in vision systems, and is the basis for further processing such as description or recognition. The goal of segmentation is to extract important features from images. Segmentation of an image can also be seen, in practice, as the classification of each image pixel to be assigned to one of the image compositions.Different segmentation methods have been presented include the dynamic system modeling based approach [3] performs pixel clustering operations in a parallel manner to speed-up the segmentation process. The cellular neural network scheme [4, 5] segments the spots by performing a number of operations such as background clean-up, grid analysis, irregular spot elimination, and intensity analysis. The morphology based approach [6] uses a series of optional steps to segment the microarray image. The combination of Markov random field based grid segmentation and active contour modeling constitutes an approach suitable for spot detection and segmentation [7]. The two-stage clustering based approach [8] is comprised of spots boundaries adjusting and intensity-based partitioning operations. The use of adaptive thresholding and statistical intensity modeling is the base for some segmentation schemes [9], whereas another approach [10] uses a seeded region growing algorithm to identify spots of different shapes and sizes. Histogram and thresholding operations were used to classify microarray image samples into either foreground (spots) or background pixels [11].One of the key steps in extracting information from a microarray image is the segmentation whose aim is to identify which pixels within an image represent which gene. This task is greatly complicated by noise within the image and a wide degree of variation in the values of the pixels belonging to a typical spot. A proposed segmentation method is presented on different noisy microarray images, which gives high accuracy. The paper is organized as follows: a brief introduction is presented in this section, section 2 presents the used materials, section 3 summarizes the proposed segmentation method applied on different cDNA microarray noisy image and section 4 discusses the results of the applied algorithm on microarray data set images. Conclusions are presented in section 5.
2. Materials
- Twenty different images have been selected from two different data sets, to test the performance of the proposed methods. They have different scanning resolutions, and different noise types, in order to study the flexibility of the proposed methods to detect spots with different sizes and features. The images are stored in TIFF files with 16-bit gray level depth. A chosen microarray image with various kinds of artifacts or noises is drawn from Princeton University Microarray (PUMA) database [12]. The data of the used images are described as follows:The first image includes thirty-two sub-grids representing acute lymphoblastic leukemia tissues (PUMA Experiment ID: 10223). The slide name is (shae082), and it is a cDNA microarray spotted by a total of 24192 genes.The second image includes thirty-two sub-grids representing acute lymphoblastic leukemia tissues (PUMA Experiment ID: 10224). The slide name is (shae083), and it is a cDNA microarray spotted by a total of 24192 genes. The third image includes thirty-two sub-grids representing acute lymphoblastic leukemia tissues (PUMA Experiment ID: 10966). The slide name is (shae086), and it is a cDNA microarray spotted by a total of 24192 genes. MATLAB [13] is used for data analysis and technical computing, as it is a high performance and powerful tool. MATLAB version is 6.5 on a windows XP platform. The P.C used has a processor: Intel Core r2 4300 CPU with 2GB of memory.
3. Methods
- Once grids have been placed [14], discrimination between areas that are considered the spot signal and areas that are considered the background signal must be carried out. The process, by which each individual cell in the grid must be selected to determine the spot signal and to estimate the background hybridization, is called segmentation. That information will be put towards a quantitative measurement at each cell. “Adaptive shape segmentation” approach is presented. Adaptive shape segmentation; seeded region growing (SRG) segmentation is a common technique that deals with different shapes in image segmentation. In SRG, the regions grow outwards from the seed points, preferentially, based on the difference between the pixel value and the running mean of values in an adjoining region. This method requires an initial point to be known, which is called the seed. After obtaining the seeds, the process is repeated simultaneously for both foreground and background regions until all the pixels are assigned to either foreground or background. Those pixels that are adjacent to a region are assigned first according to its intensity. “Fig. 1” shows the steps of applying the method.
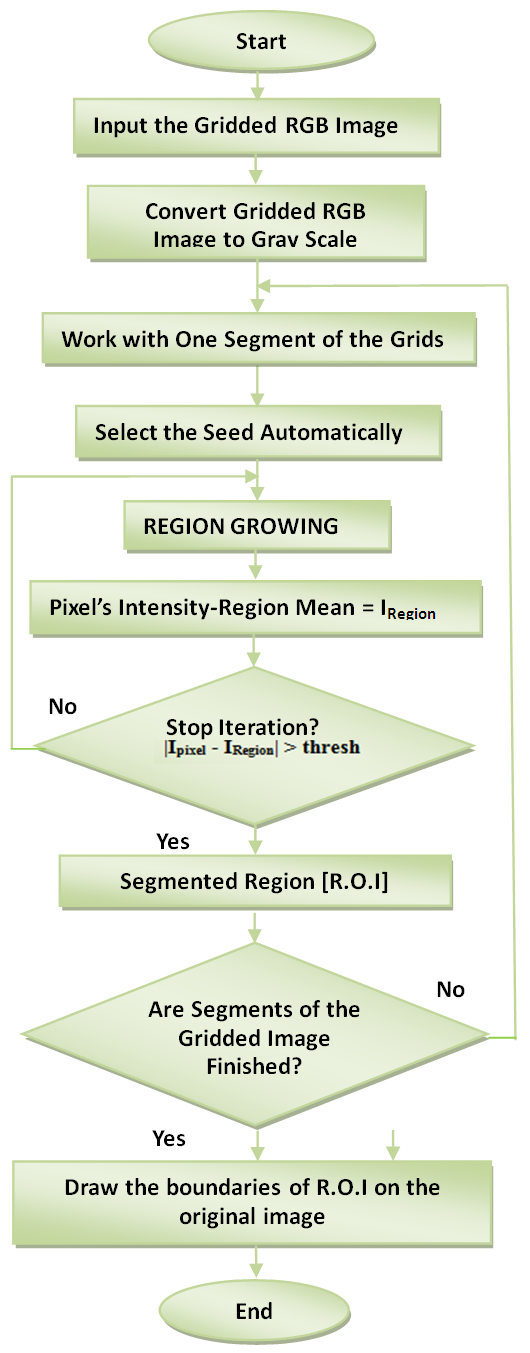 | Figure 1. Flowchart of adaptive shape segmentation |
4. Results and Discussions
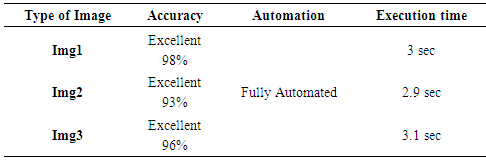
- The presented segmentation method is tested on a number of noisy microarray images drawn from Princeton University Microarray (PUMA) database. Depending on the degree of noise and how the spots are expressed, three noisy microarray images are used. The following figures from “Fig. 2” to “Fig. 4” show results of applying the proposed method on three samples of images having different qualities.
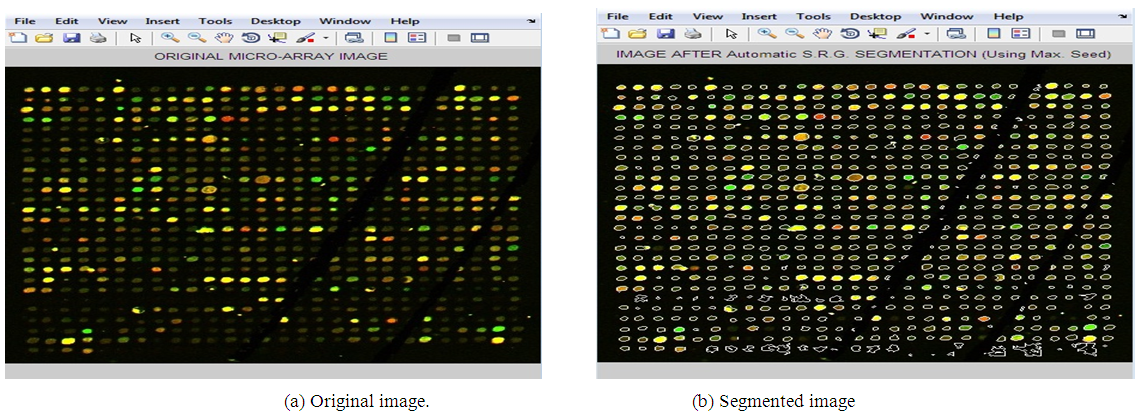 | Figure 2. Applying Adaptive Shape Segmentation Automatically to Image 10223Using a Maximum Seed. (Img1: PUMA Experiment ID: 10223, Sub-array 4th row, 2nd column.) |
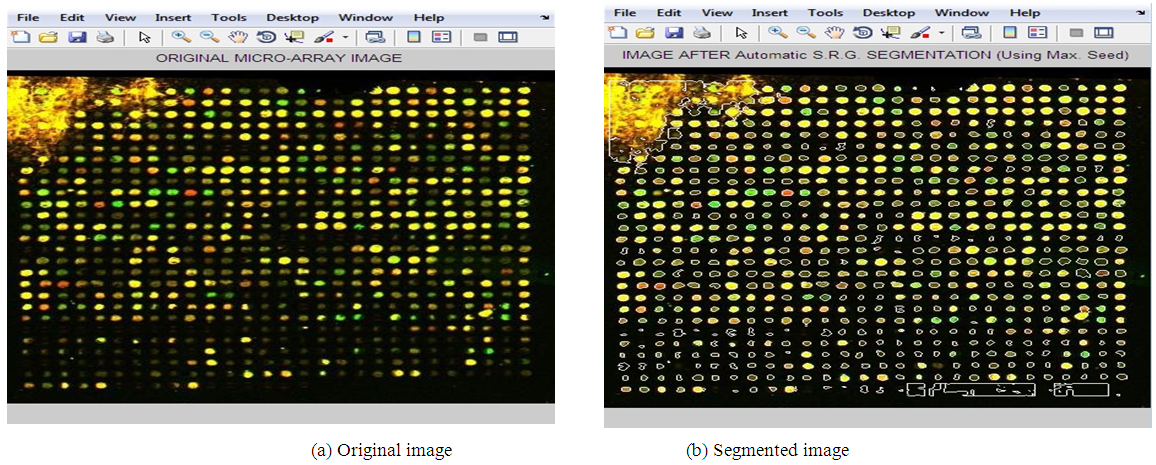 | Figure 3. Applying Adaptive Shape Segmentation Automatically to Image 10224 Using a Maximum Seed. (Img2: PUMA Experiment ID: 10224, Sub-array 1st row, 4th column) |
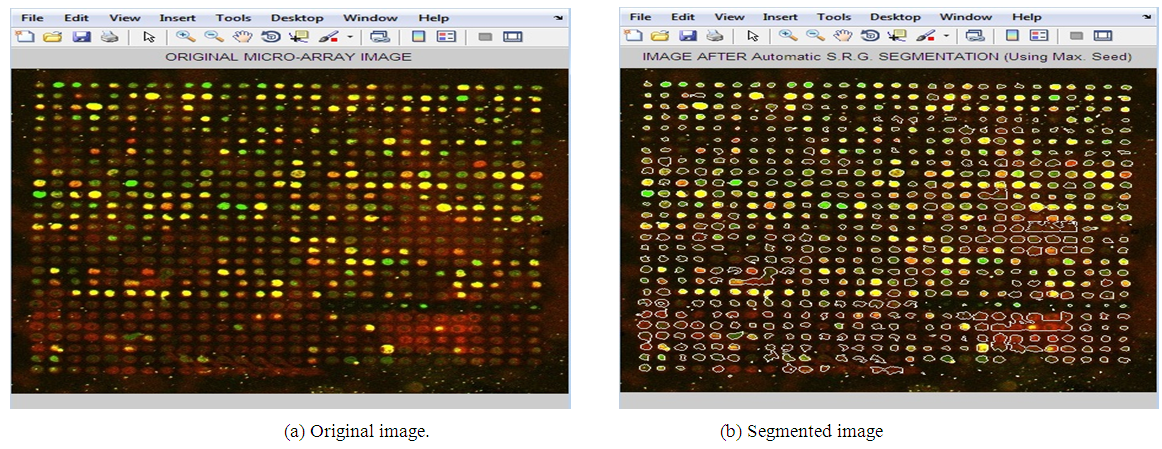 | Figure 4. Applying Adaptive Shape Segmentation Automatically to Image 10966 Using a Maximum Seed. (Img3: PUMA Experiment ID: 10966, Sub-array 8th row, 4th column) |
|
5. Conclusions and Future Work
- The Microarray technology has a great potential in obtaining a deep understanding of the functional organization of cells. The emergence of this technology allows the researchers to tackle difficult problems and reveal promising solutions in many fields. In this respect, our research based heavily on the requirement for a reliable yet time efficient automated method. One of the key steps in extracting information from a microarray image is the segmentation whose aim is to identify which pixels within an image represent which gene. This task is greatly complicated by noise within the image and a wide degree of variation in the values of the pixels belonging to a typical spot. This work emphasizes the impact of microarray image segmentation. In the segmentation stage, efficient segmentation method has been presented; “adaptive shape segmentation” method. When the proposed method inspected, it was found clearly that “adaptive shape segmentation” gives fine results using a maximum seed.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML