-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Clinical Medicine and Diagnostics
p-ISSN: 2163-1433 e-ISSN: 2163-1441
2018; 8(4): 59-62
doi:10.5923/j.cmd.20180804.01

Association of BCR-ABL Transcripts Variants and Haematological Features among Philadelphia Chromosome-Positive CML Sudanese Patients
Abdalla A. A. Elnour1, Mahdi H. A. Abdalla2
1Department of Basic Science, Faculty of Medicine, King Abdul-Aziz University, Rabigh, Saudi Arabia
2Department of Haematology, Faculty of Medical Laboratory Sciences, Omdurman Ahlia University, Sudan
Correspondence to: Mahdi H. A. Abdalla, Department of Haematology, Faculty of Medical Laboratory Sciences, Omdurman Ahlia University, Sudan.
| Email: |  |
Copyright © 2018 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Background: The types of the fusion gene in CML are thought to be related to the disease clinical course and outcome. The hematological characteristics of the common breakpoint cluster region-Abelson (BCR-ABL) transcript variants p210 (e13a2, e14a2) and p190 (e1a2) has been reported from CML Sudanese patients, but there were variation and limitation in the published data in Sudan. Aims and Objectives: This study aimed to highlight the association of the hematological characteristics and BCR-ABL transcript variants (p210 and p190) among Ph'-positive CML Sudanese patients. Materials and Methods: A total of One hundred confirmed cases for Philadelphia chromosome-positive CML Sudanese patients were enrolled in this study and a quantitative real-time – polymerase chain reaction technique (qRT-PCR) was used to detect BCR-ABL transcript variants p210 (Major-BCR), and p190 (Minor-BCR) in CML patients, Complete blood cells count were performed by an automated cell counter (XP-300 Sysmex). Result: 23% of CML patients were thrombocytopenic (Plt <150×109/l), while 19% were thrombocythemic (Plt >450×109/L). 69% Of CML patients were anemic (Hb <120g/L). The mean Plt count of b2a2 was 417±286×109/L, the mean Plt count of b3a2 was 308±210×109/L and the mean Plt count of co-expression group (b3a2/e1a2) was 539±431×109/L with P-value of 0.035. Conclusion: There was a significant correlation (P-value < 0.05) in the mean of Platelets count between, b3a2 and co-expression (b3a2/e1a2) BCR-ABL transcripts variants, P-value 0.014.
Keywords: CML Sudanese patients, Philadelphia chromosome, BCR-ABL
Cite this paper: Abdalla A. A. Elnour, Mahdi H. A. Abdalla, Association of BCR-ABL Transcripts Variants and Haematological Features among Philadelphia Chromosome-Positive CML Sudanese Patients, Clinical Medicine and Diagnostics, Vol. 8 No. 4, 2018, pp. 59-62. doi: 10.5923/j.cmd.20180804.01.
Article Outline
1. Introduction
- Chronic myeloid leukemia (CML), also known as chronic myelogenous leukemia is a myeloproliferative neoplasm [1]. CML characterized by an abnormal chromosome 22 (truncated) that results from reciprocal translocations between chromosomes 9 and 22 [t (9; 22) (q34; q11)], which called Philadelphia chromosome (Ph') [1]. As a result of this reciprocal translocation, the BCR-ABL oncogene was created, which was translated into a protein with constitutive tyrosine kinase activity [2].The BCR-ABL genomic breakpoint location is highly variable [3]. According to the breakpoints in the BCR gene, three BCR-ABL genes can be formed: The first typical BCR-ABL gene that is seen in more than 99% of Ph'-positive CML patients is derived from a disruption of the major breakpoint cluster region (M-bcr) either between b2 and b3 or b3 and b4 which results in (b2a2) or (b3a2) transcripts. This result in translation of a 210 kDa fusion protein designated as (p210 BCR-ABL). The second breakpoint in the BCR gene has been identified in Ph'-positive ALL and in occasional cases of CML and is located in intron 1 within the minor breakpoint cluster region (m-bcr). Consequently, only BCR exon 1 (e1) is joined to ABL exon 2 (e1a2) transcript, the translation results in (p190 BCR-ABL) protein. The third break-point is located in the micro break-point cluster region (μ-bcr) between exon 19 and exon 20 and results in an (e19a2) BCR-ABL transcript and a (p230 BCR-ABL) protein. Another rare transcript (b2a3, b3a3) that occurs within the (M-bcr) region can also be seen in CML. Co-expression of the transcripts in the (M-bcr) or the (m-bcr) with one of (M-bcr) have also been reported. Breakpoints in the ABL gene are relatively consistent, typically in the intron before exon 2 [4, 5].The types of the fusion gene in CML are thought to be related to the disease clinical course and outcome. Researchers have been, however, unsuccessful in locating any significant correlation [5, 6].
2. Materials and Methods
- One hundred Ph'-positive CML patients were enrolled in this study. Following informed consent, 3 ml of peripheral blood samples were collected in EDTA from each patient. Hematological and molecular analyses were performed at RICK and Alzahrawi Medical laboratories, Khartoum, Sudan.
2.1. RNA Extraction
- Leukocytes were prepared from peripheral blood samples after the addition of red blood cells lysis buffer (0.1mM EDTA and 1mM KHCO3, 150mM NH4Cl) pH 7.3. Total RNA was extracted from mononuclear cells by the TRIzol reagent. Extracted RNA integrity was determined by gel electrophoresis (agarose).
2.2. cDNA Synthesis
- For cDNA synthesis, 5μl of total RNA was first incubated with 9.5 ml of RNAase free distilled water at 70°C for 10 minutes. Cooled on ice and reversely transcribed in a reaction mixture containing (Reverse Transcriptase (RT) buffer: 20 mM Tris HCl, 50 mM KCl, pH 8.3; 5mM MgCl2, 10 mM DTT, 5mM random hexamers, 20 units RNAase, 10 units RT enzyme, 1mM dNTP and H2O to a total volume of 20 μl. At 42°C for 60 minutes. RT enzyme was denatured by incubating the reaction at 99°C for 5 minutes.
2.3. Quantitative Real-Time – Polymerase Chain Reaction (qRT-PCR)
- Primers and probe sequences were obtained from (Eurofins genomics) company as follows: (BCR2) b2 sense: TGCAGA TGC TGA CCA ACT CG; (BCR3) b3 sense: CGT CCA CTC AGC CAC AT; and a2 (ABL) antisense: TCCAAC GAG CGG CTT CAC. TaqMan probe for e14a2 and e13a2 was: CAG TAG CAT CTG ACT TTG AGC CTC AGG GTC T, which is derived from ABL exon 2 and lies within the fusion region of the b2a2 and b3a2. The ABL Primers and probe were: ABL antisense, GGC CAC AAA ATC ATA CAG TGC A; ABL sense, GTC TGA GTG AAG CCG CTC GT; and TaqMan probe, TGG ACC CAG TGA AAA TGA CCC CAACC. Sequences were contained in ABL exon 2. Reaction mixtures of 25 μl contained MgCl2 5 mM, TaqMan buffer A with the ROX dye as the passive reference 12.5 μl. 400μM dUTP, 200μMdATP, dCTP, dGTP, AmpliTaq Gold DNA polymerase 1.25 U, Amp Erase Uracil N-glycosylase (UNG) 0.5 U, forward and reverse primers 300 nM, specific TaqMan probe 200 nM, and cDNA 6μl (1:3). Dilution Mixture was incubated at 50°C for 2 minutes and 95°C for 10 minutes, with 50 amplification cycles of 15 seconds at 95°C and 60 seconds at 65°C. Agarose gel was used to determine the P210 and P190 transcript variants by differences in fragment size of PCR product.
2.4. Ethical Consideration
- The participants were informed by simple language about the aim of the research, the disease and the benefit from the study. Patients consent were taken if they agreed to participate. The blood samples were collected under aseptic technique by a professional technician to protect the participants from infectious hazards.
3. Results
3.1. Demographic Data
- A total of One hundred confirmed cases of Ph'-positive CML Sudanese patients' samples were examined by qRT-PCR for BCR-ABL transcripts variants. 59% (n=59) were males while 41% (n=41) were females. Males: females' ratio were 1.5:1 with P-value 0.072. Mean age of males was 46.56±8.8 years while the mean age of females was 46.51±9.5 years.
3.2. Hematological Profiles
- The means of the haematological values were as follow: Platelets count was 328.8×109/L±240, total leukocyte count was 84.5×109/L±103, Haemoglobin level was 110.4g/L±21, basophils was 1.29% ±0.71 and blast 1.53% ±0.83. 23% of CML patients were thrombocytopenic (Plt <150×109/l), while 19% were thrombocythemic (Plt >450×109/L). 69% Of CML patients were anemic (Hb <120g/L). No patients had myeloblast more than 10% in peripheral blood. 17% of CML patients were basophilic (basophil >1%).
3.3. Association between Transcripts Types in Hematological Profiles
- The mean Plt count of b2a2 was 417±286×109/L, the mean Plt count of b3a2 was 308±210×109/L and the mean Plt count of co-expression group (b3a2/e1a2) was 539±431×109/L with P-value of 0.035 as shown in figure (1), and other as shown in Table (1).
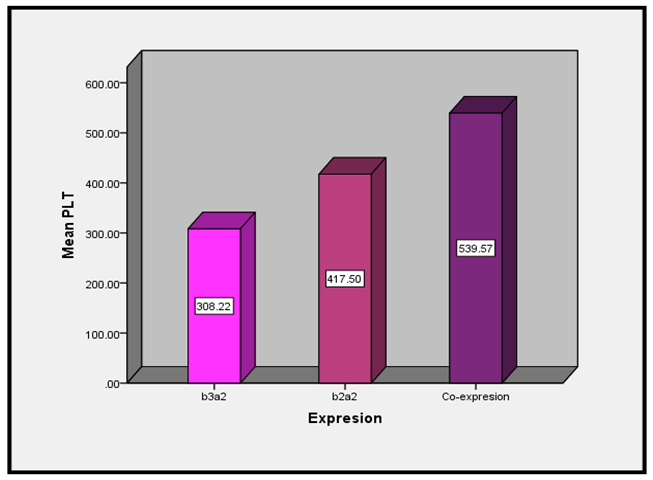 | Figure (1). Association between Transcripts and platelets count |
|
4. Discussion
- Our study detected that: males: females' ratio was 1.5:1, this result was comparable with international, Arabian and even local previous studies [7-9]. Our research detected that: the mean age was 46±9 years, while the previous report identified semi-comparable mean age 50±14 years [8], however, other report detected younger mean age 39±16 years [10]. This study detected some diverse in resemblance and contradictory in hematological profiles between previous studies. Our study detected that: mean of platelets was 328.8±240×109/L; this result come to agree with other previous local report showed that: the mean platelets was 369±238×109/L [7]. However, our result disagreed with other report showed, the mean of Plt was 615±406×109/L [8]. Also, Our study detected that: the mean of Haemoglobin was 110±21 g/L, this result come to agree with other previous report showed, the mean of Hb was 111±27g/L [8]. Also, our study detected that: the mean of TLC was 84.5±103×109/L, this result slightly lower than the previous local study showed, the mean of TLC was 149±92×109/L [7], and lower than the previous international research showed: the mean of TLC was 125±90×109/L [8].Our study detected that: 69% (n=69) of patients were anemic, which was come close to the previous research showed, 69.8% (n=58) were anemic [9]. Also, this study showed 23% (n=23) of patients were thrombocytopenic, while 19% (n=19) have thrombocytosis, which was agreed with the previous study showed, 9% had thrombocytosis. However, our result disagreed with previous research showed no patients had thrombocytopenia [10].Our study detected that co-expression of (b3a2/e1a2) BCR-ABL transcripts variants is associated with high Platelets count (P-value 0.014), so there is a possible role of e1a2 transcripts in this association, this suggestion supports previous reports that described an association between p190 and rapid disease prognosis [12]. This study showed that no significant difference between BCR-ABL transcripts in the other hematological parameters, which came to agree with other recent researches [9, 12]. The disagreement between previous studies and our study could be probably due to genetic factors, sample size or other confounding factors. Also, the differences between local study probably because Sudan is vas countries with several types of ethnic groups [7].
5. Conclusions
- This study concluded that mean Plt count of b2a2 was 417±286×109/L, the mean Plt count of b3a2 was 308±210×109/L and the mean Plt count of co-expression group (b3a2/e1a2) was 539±431×109/L. Co-expression of (b3a2/e1a2) BCR-ABL transcripts variants is associated with high Platelets count.
ACKNOWLEDGMENTS
- We would like to express our sincere appreciation and gratitude to Alzahrawi Medical Laboratories Staff.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML