-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Clinical Medicine and Diagnostics
p-ISSN: 2163-1433 e-ISSN: 2163-1441
2016; 6(2): 38-43
doi:10.5923/j.cmd.20160602.03

Factors of Risk Survey of the Nosocomial Infection in the Hospital Inert Environment
Rakotozafy J. C. R.1, Raherimandimby H.2, Rasoloherimampiononiaina M. R.3, Rakotoarimanana R.2, Randriamamonjy F.2, Tanamasoandro R.3, Solofomalala G. D.3, Rabarijaona M.3
1Laboratory of Molecular Biology, Faculty of Sciences – University of Fianarantsoa, Madagascar
2Laboratory of Biomedical Analysis, Center Hospital University of Fianarantsoa, Madagascar
3Center Hospital University of Fianarantsoa, Madagascar
Correspondence to: Rakotozafy J. C. R., Laboratory of Molecular Biology, Faculty of Sciences – University of Fianarantsoa, Madagascar.
| Email: |  |
Copyright © 2016 Scientific & Academic Publishing. All Rights Reserved.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

In this survey, we analyzed by means of the inert environment withdrawals at the Resuscitation service, Neonatalogy and Operative Block in the Center Hospital University of Fianarantsoa (Madagascar). The bacteria identification has been done by biomedical laboratory usual technique. The results showed that different bacterial species contaminate the inert environment of these three services. Two bacterial species: Staphylococcus aureus and Escherichia coli are frequently isolated on these different samples. The antibiogram on disk method showed the presence of oxacillin and vancomycin-resistant Staphylococcus aureus; vancomycin-sensitive Staphylococcus aureus andoxacillin-resistant Staphylococcus aureus. For Escherichia coli, there are species cefoxitin and augmentin-sensitive Escherichia coli and cefoxitin and augmentin-resistant Escherichia coli. These results shows that the risks of nosocomial infections from the inert environment at the hospital is raised. The future perspective of this survey is to identify these two bacterial species isolated by molecular technique and to look for the new antibacterial products.
Keywords: Antibiogram, Environment, Escherichia coli, Fianarantsoa, Madagascar, Staphylococcus aureus
Cite this paper: Rakotozafy J. C. R., Raherimandimby H., Rasoloherimampiononiaina M. R., Rakotoarimanana R., Randriamamonjy F., Tanamasoandro R., Solofomalala G. D., Rabarijaona M., Factors of Risk Survey of the Nosocomial Infection in the Hospital Inert Environment, Clinical Medicine and Diagnostics, Vol. 6 No. 2, 2016, pp. 38-43. doi: 10.5923/j.cmd.20160602.03.
Article Outline
1. Introduction
- The nosocomial infections constitute a real problem of public health. 5 to 10% of the hospitalized patients incur a nosocomial infection during their hospitable stay [1].The nosocomial infections is defined like infections appearing throughout a hospitalization [2] [3]. It can concern all types of infectious agents, but they are most frequently bacterial, and more occasionally viral, fungal or parasitic [2].Many factors contribute to increase the risks like patient, cares, interventions, maternal-fetal infection and the inert environment contaminated [4]. These infections are the main reason of death neonatal in Africa, valued to 39% [5] [6].The World Health Organization (WHO), published in April 30, 2014 a report on antibiotics resistance in every region of the world.Indeed, inert environment’s ecology bacterial has been done at the hospital. This survey has objective to identify the bacterial contamination to be able to take an adequate and perennial measure to the preventive and curative struggle.
2. Material and Methods
2.1. Sites of Study
- The study has been achieved to the Center Hospital University of Fianarantsoa (Madagascar) in the Operative Block, Resuscitation and Neonatalogy Services, in 2014. The choice of these three services is based on the bacterial contamination high risk.
2.2. Withdrawal of the Environment
- Different materials touched by the health staff, patients and their accompanists have been swabbed with sterile dry swabs. The withdrawals have been done between 2 to 3 pm when every service daily activity is minimal during 15 days a month.The appropriated samples have been sent to the laboratory for bacteria isolation and identification.
2.3. Bacteria Identification
- Staphylococcus aureus, Streptococcus spp, Pseudomonas aeruginosa, Escherichia coli, and Salmonella spp are the most frequently isolated and identified pathogenic bacterial to the hospitalized patients. Indeed, the study was focused on these kinds of bacteria. All withdrawals don't possess one of these bacterial species are excluded.Bacteria identification and antibiogram have been achieved by biomedical laboratory usual technique [7] [8] [9] [10].
2.3.1. Media Culture
- A preliminary identification by Gram coloration (Kit Gram-Nicolle RAL®) has been achieved for every sample, to be able to classify the bacteria in cocci and bacillus group.According to the results of Gram methods, different culture medium are used to isolate the bacteria:- Staphylococcus aureus: on salty media (Chapman, BioRad®), some yellow colonies were looked for after 24 hours of 37°C incubation. - Streptococcus spp: on trypticase soy media (BioRad®) to 5% of sheep blood, small colonies surrounded a halo were searched after 24 hours and 48 hours of 37°C incubation under 5% of CO2.Three media have been used for bacillus Gram negative:- Pseudomonas aeruginosa: the presence of colony producing bluish fluorescent pigments was searched on culture medium (KING A, BioRad®) after 48 hours of 37°C incubation,- Escherichia coli:, the sample is sowed on media (UriSelect, BioRad®) to isolate Escherichia coli. Purple colonies were looked for on culture medium after 48 hours of 37°C incubation.- Salmonella spp: black colonies were searched on Salmonella Shigella agar (SS Agar, BioRad®) media after 48 hours of 37°C incubation.
2.3.2. Biochemical Test (API-System)
- API System (bioMérieux®) is a standardized system for genera bacteria identification, which uses miniaturized biochemical tests and a specially adapted database. The complete list of those bacteria that is possible to identify with this system can be found in the Identification Table at the end of this package insert.The API System strip consists of 20 microtubes containing dehydrated substrates. These microtubes are inoculated with a bacterial suspension, prepared in API System Medium that reconstitutes the tests. During incubation, metabolism produces color changes that are either spontaneous or revealed by the addition of reagents.The reactions are read according to the Reading Table and the identification is obtained by referring to the Analytical Profile Index or using the identification software [11].
2.4. Antibacterial Disk Susceptibility Tests
- Antibiotic susceptibility testing by a standardized single disk method have been used [7] [8] [9] [10]. Four milliliters of this bacterial suspension (1-3 X 104 bacteria/ml) have been spread uniformly on the Mueller Hinton agar media [BioRad®]. Afterwards, the different antibiotics disks (Cypress Diagnostics®) have been deposited on media. After 48 hours of 37°C incubation (under an atmosphere enriched of 5% CO2 for Streptococcus spp.), the inhibition area diameter has been measured (Figure 1). Every antibiotic has been tested 5 times. The results are expressed in millimeter (mm).
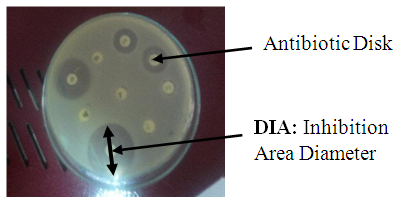 | Figure 1. Inhibition Area Diameter measure technique |
2.5. Statistical Analysis
- The statistical analysis of the qualitative and quantitative variables was either of mono-varied type, or of multivariate type using a software Epinfo version 6. The Student test was used for the averages and the variances comparison and the test of Chi-2 for the comparison between the rates and a significant value if p < 0. 05.
3. Results and discussions
3.1. Operative Block
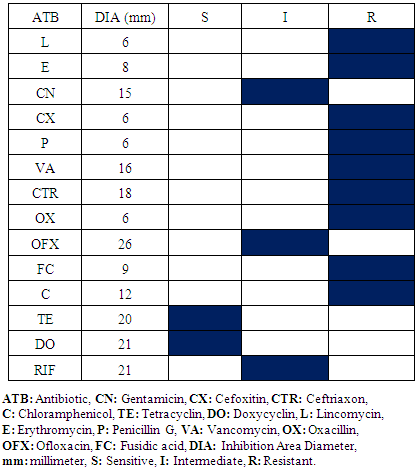
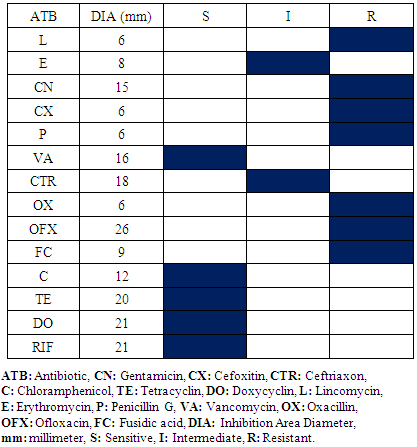
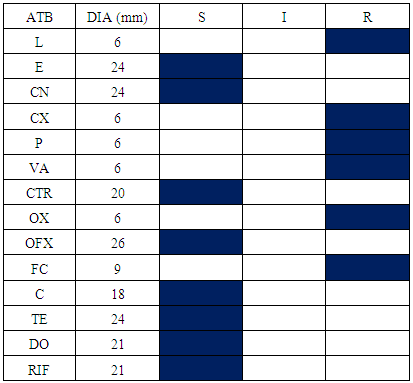
- The samples appropriated results within the Operative Block show the presence of Staphylococcus aureus and Escherichia coli. The difference of antibiogram results is remarkable within Staphylococcus aureus compared with oxacillin and vancomycin activity and Escherichia coli within ceftriaxon and augmentin. Consequently, there are oxacillin and vancomycin- resistant Staphylococcus aureus; vancomycin-sensitive Staphylococcus aureus and oxacillin-resistant Staphylococcus aureus (Table 1, 2). Otherwise, cefoxitin and augmentin-sensitive Escherichia coli and cefoxitin and augmentin-resistant Escherichia coli exist (Table 3, 4). Antibiogram results show that all Staphylococcus aureus were sensitive in doxycyclin and tetracyclin while all Escherichia coli are sensitive in imipenem and gentamicin.
|
|
|
|
3.2. Resuscitation Service
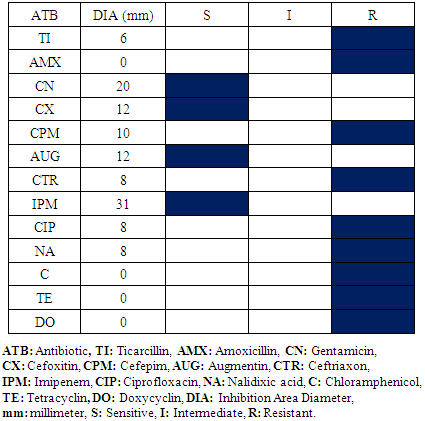
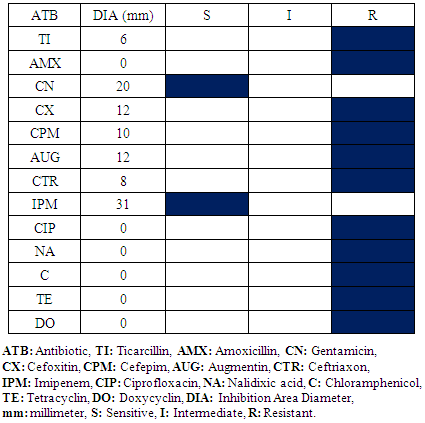
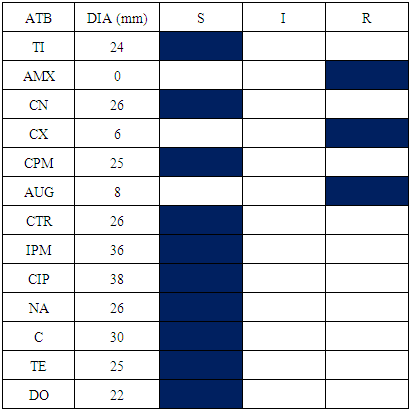
- Four bacterial species have been isolated and identified in this service: Staphylococcus aureus, Escherichia coli, Staphylococcus epidermis, Staphylococcus saprophyticus, Klebsiella spp and Proteus spp.Every bacterial species antibiogram results are represented in table 5, 6, 7 and 8. For Staphylococcus aureus and Escherichia coli, the results are identical to the Operative Block. The difference is not meaningful.
|
|
|
|
3.3. Neonatalogy Service
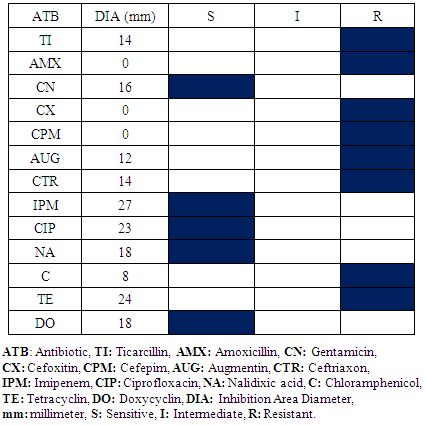
- All bacterial species isolated in Operative Block and Resuscitation room are also isolated to Neonatalogy Service except Klebsiella spp. No meaningful difference is observed to the antibiogram results within the Operative Block and the Resuscitation room vis-a-vis Staphylococcus aureus and Escherichia coli.It is noted that Enterobacter spp. and Pseudomonas aeruginosa are isolated in those three services environment (Table 9, 10).
|
|
4. Conclusions
- This survey shows the bacteria multi resistant presence on the inert environment at the hospital. Indeed, nosocomial infections risk is raised. Even that the probable sources of nosocomial infection to patients in these three services are not studied, these results permit to take preventive and curative adequate measures at the hospital as creation of surveillance service assures cleanings; the cleanliness or even environment sterilization, local and materials used. These results also allow the clinicians to choose one among the sensitive antibiotics according the laboratory analyses results. For a perennial struggle, sources infection research; the responsible agents’ identification by molecular biology method (PCR) and new products antimicrobials research prove to be necessary.
ACKNOWLEDGMENTS
- This work was made possible with the “Fondation Mérieux – Lyon (France) and Center Hospital University of Fianarantsoa (Madagascar).
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML