-
Paper Information
- Next Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Clinical Medicine and Diagnostics
p-ISSN: 2163-1433 e-ISSN: 2163-1441
2015; 5(2): 22-25
doi:10.5923/j.cmd.20150502.02
NAD (P) H Quinine Oxidoreductase (NQO1) as a Risk Modifier of Susceptibility to Chronic Myeloid Leukaemia in Sudan
Hiba A. E. Karrar1, Mahdi H. A. Abdalla2
1Department of Haematology, Faculty of Medical Laboratory Sciences, Alneelain University, Sudan
2Department of Haematology, Faculty of Medical Laboratory Sciences, Omdurman Ahlia University, Sudan
Correspondence to: Mahdi H. A. Abdalla, Department of Haematology, Faculty of Medical Laboratory Sciences, Omdurman Ahlia University, Sudan.
| Email: |  |
Copyright © 2015 Scientific & Academic Publishing. All Rights Reserved.
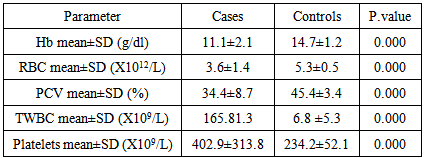
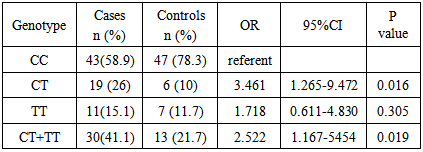
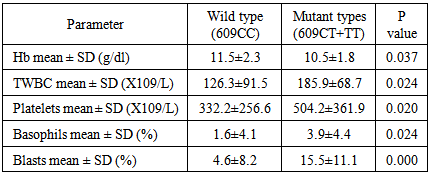
CML is the most prevalent haematological cancer among Sudanese population. Previous studies reported an association between NQO1 polymorphism and leukaemia, however these studies showed differences in the occurrence and frequency of this relationship. This study aimed to examine the association of NQO1 C609T polymorphism with the risk of CML and the clinical outcome among Philadelphia positive CML patients in Sudan. The study included 73 newly diagnosed Philadelphia positive CML patients, their NQO1 C609T genotypes (detected by PCR/RFLP) and haematological characteristics (determined by Sysmex KX-21N) were determined and compared with 60 age and sex matched normal subjects as control. When the NQO1 609CC genotype was defined as the reference, a 3.5-fold increased risk of CML for those carrying NQO1 609CT (heterozygous) genotype was observed (OR 3.461, P value 0.016). The frequency of NQO1 609 TT (homozygous) genotype was higher among CML patients with a 1.7 folds than control group, but with no statistical significance (OR 1.718, P value 0.305). The frequency of the NQO1 609CT and 609 TT genotypes combined together (mutant types) was significantly higher among CML patients with a 2.5- fold increased risk when compared with the controls (OR 2.522, P value 0.019). We observed a statistically significant reduction in the mean Hb level in patients with mutant genotypes than in wild type patients (p value 0.037), WBCs count, platelet count, basophiles count and blasts count were significantly higher in patients with mutant type when compared to those with wild type (p value 0.024, 0.020, 0.024 and 0.000) respectively. In conclusion, our results indicate that NQO1 C609T mutant genotypes, with low enzymatic activity, are associated with increased risk of CML and worse clinical outcome.
Keywords: NQO1 polymorphism, CML, Sudan
Cite this paper: Hiba A. E. Karrar, Mahdi H. A. Abdalla, NAD (P) H Quinine Oxidoreductase (NQO1) as a Risk Modifier of Susceptibility to Chronic Myeloid Leukaemia in Sudan, Clinical Medicine and Diagnostics, Vol. 5 No. 2, 2015, pp. 22-25. doi: 10.5923/j.cmd.20150502.02.
Article Outline
1. Introduction
- Chronic myeloid leukaemia (CML) (also known as chronic myelogenous leukemia, chronic Granulocytic leukemia and chronic myleocytic leukemia) is a clonal myeloproliferative disorder of pluripotent stem cell [1,2]. The disease accounts for around 15% of leukaemia with an incidence of 1-2 cases/100,000 population [3]. CML is the most prevalent haematological cancer among Sudanese male, and the second prevalent cancer, after breast cancer, among Sudanese female [4].The median age, at diagnosis is 45-55 years, but all age groups including children can be affected [5]. CML has three clinical stages: a chronic phase that is characterized by an indolent onset without symptoms in 50% of patients. This is followed invariably by an accelerated phase that is characterized by basophilia, increased peripheral blood blasts and promyelocytes and is refractory to treatment. In about 75% of patients, the accelerated-phase disease is followed by a blastic phase, which resembles acute leukaemia and causes death within 3 to 6 months [6].CML was the first neoplasm to be associated with a specific chromosomal rearrangement, the Philadelphia chromosome, that results from a reciprocal translocations between chromosomes 9 and 22 [t (9; 22) (q34; q11)]. This translocation transposes the c-abl proto-oncogene from chromosome 9 to the breakpoint cluster region (bcr) gene on chromosome 22, and this new hybrid bcr-abl oncogene encodes a constitutively active tyrosine-kinase that promotes the growth advantage of leukemia cells through enhanced proliferation and reduced apoptosis, and generating genomic instability [7]. Although the clinical and biological aspects are well documented, little is known about individual susceptibility to CML. Polymorphic variants of several genes, diet, environmental exposure to carcinogens and individual immune system’s characteristics are potential factors that increase predisposition to leukemia [8] Finding of resent studies suggest that genetic variants may affect and increase risk of CML, and among them are methylenetetrahydrofolate reductase (MTHFR) [9, 10], glutathione S-transferases (GSTs) [11, 12], and NAD(P)H: Quinone Oxidoreductase (NQO1) [13].(NQO1) gene is located in the long arm of chromosome 16 (16q22.1), it expands approximately 20 kb with 6 exons and 5 entrons that code for NQO1 protein, a flavoenzyme mainly cytosolic enzyme formed of 273 amino acid residues, that plays necessary role in the protection against exogenous and endogenous quinone by catalyzing two and four electron reduction of these substrates, such as hydroquinone. [14-16]. NQO1 enzyme has many functions that include protection of the cells from oxidative damage, scavenging of superoxide, stabilization of p53 and other tumor suppressors and detoxification of quinone and their derivatives. [15] The polymorphism occurs at exon 6 at nucleotide 609 (C-T) in the human NQO1 gene results in a proline to serine substitution at position 187 in the amino acid structure of the NQO1 protein [17]. NQO1 enzyme activity is normal in individuals with 2 wild-type alleles (NQO1 609CC). It is variably reduced in individuals who are heterozygotes for the polymorphism (NQO1 609CT) [18]. NQO1 enzyme activity is absent in those who are homozygous for the point mutation (NQO1 609TT) [19].Previous studies reported an association between NQO1 polymorphism and leukaemia; however these studies showed differences in the occurrence and frequency of this relationship. The aim of this study was to examine the association of NQO1 C609T polymorphism with the risk of CML and the clinical outcome among Philadelphia positive CML patients in Sudan.
2. Materials and Methods
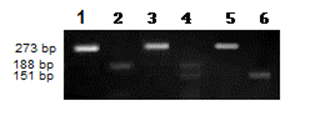
- Following informed consent 133 individuals were enrolled, 73 newly diagnosed Philadelphia positive CML patients (Diagnosis based on the haematological features and the detection of BCR-ABL transcripts) who were attended radiation and isotopes center of Khartoum (RICK); and 60 apparently healthy subjects as controls.Two ml of EDTA anticoagulated blood was collected from each subject for haematological and molecular analysis. Laboratory investigations were performed at the department of haematology, faculty of medical laboratory sciences, Alneelain University, Sudan. Blood cell count was performed by automated cell counter (Sysmex KX-21N). DNA was extracted by DNA extraction kit (analytik Jena Biometra, Germany), according to manufacturer’s instructions. NQO1 fragment Was Amplified using the forward primer: 5`-AGTGGCATTCTGCATTTCTGTG-3` and reverse primer:5`-GATGGACTTGCCCAAGTGATG-3`. The amplification was carried out in thermo-cycler (Techne) with initial denaturation step for 8 minute at 95°C Followed by 35 Cycles consisting of 3 steps: Denaturing step at 94°C for 30 second, Annealing step at 56°C For 1 minute and extension steps at 72°C for 40 minute with final Extension step at 72°C for 10 Minutes.The PCR reactions was performed in a final volume of 20 µl containing (4 μl premixed ready to use 5x FIREPol master mix (Solis BioDyne,Russian), 12.0μl DNAase free DW, 3 μl genomic DNA and 0.5 μl from each primer). The amplified fragment was digested with 10 U Hinf1 endonuclease (New England Bio lab, UK) over night and was visualized on agarose gel electrophoresis.Statistical analysis was performed using statistical package for social science (SPSS) software. Evaluation of patient´s data was performed using t-test. Comparison of frequency distribution between groups was made by means of the X2 test. All tests are two-sided and P-value less than 0.05 have been considered as statistically significant. Crude odds ratios (OR) were also calculated and given with 95% confidence intervals (CI).
3. Results
- The male: female ratio was 1.5 and the median Age was 43 year, with minimum Age of 8 and maximum of 80 years. All patients were tested for the blood cell counts and NQO1 Polymorphism.
|
|
|
4. Discussion
- Genetic polymorphisms of various kinds of genes have been recently proved to have important roles in the genesis of human malignancies [20]. Several studies have reported that individuals with NQO1 C609T mutant genotypes are at increased risk of leukemia. We examined the association between NQO1 C609T polymorphism and the risk of CML. Our study included 73 CML patients, their NQO1 C609T genotype frequencies and haematological characteristics were determined and compared with 60 age and sex matched normal subjects as control.To determine if there is a statistically significant increase risk of CML development according to the NQO1 genotypes, we conducted logistic regression analysis, our study showed a statistically significant association between NQO1 C609T polymorphisms and the risk of CML. The frequency of the NQO1 609CC genotype was higher among controls (45.0%) when compared to CML patients (35.2%). When odds ratios were calculated for the overall group, we observed a 3.5-fold increased risk of CML for those carrying NQO1 609CT (heterozygous) genotype (OR 3.461, P value 0.016). The frequency of NQO1 609 TT (homozygous) genotype was higher among CML patients (15.1%) than control group (11.7%), but with no statistical significance (OR 1.718, P value 0.305). The frequency of the NQO1 609CT and 609 TT genotypes combined together (mutant types) was significantly higher among CML patients (41.1%) when compared with the controls (21.7%), with a 2.5-fold increased risk of CML (OR 2.522, Pvalue 0.019). Similar findings had previously been reported [20].When comparing the haematological values between CML patients with NQO1 C609T wild type (CC) and those with mutant types (CT and TT genotypes combined together), we observed a statistically significant reduction in the mean Hb level in patients with mutant genotypes than in wild type patients, WBCs count, platelet count, basophiles count and blasts count were significantly higher in patients with mutant type when compared to those with the wild type. Higher, platelets count, basophils count and blast count are associated with worse clinical outcome in CML [21, 22], thus, in our study group, clinical outcome was worse among CML patients with NQO1 C609T mutant types (CT and TT genotypes combined together) than among patients with wild type (CC). Reduced detoxifying power for toxic quinone and free radicals and/or the decreased stability of p53 resulting from the NQO1 inactivating polymorphism may influence the susceptibility to CML. However, further investigation needs to verify this hypothesis and to understand the mechanism.
5. Conclusions
- In conclusion, we examined the association between NQO1 C609T polymorphism and the risk of CML. NQO1 C609T genotypes and haematological characteristics of 73 CML patients were determined and compared with 60 age and sex matched normal subjects as control. Our results indicate that NQO1 C609T mutant genotypes with low enzymatic activity are associated with increased risk of CML and worse clinical outcome.
ACKNOWLEDGEMENTS
- Special thanks to the Staff of Haematology Department, Faculty of Medical Laboratory Sciences, Alneelain University and the staff radiation and isotopes center of Khartoum.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML