-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Clinical Medicine and Diagnostics
p-ISSN: 2163-1433 e-ISSN: 2163-1441
2014; 4(4): 71-77
doi:10.5923/j.cmd.20140404.03
Association of Interleukin 13 +2044G/A Polymorphism with Bronchial Asthma Development, Severity and Immunoglobulin E levels in an Egyptian Population
Mohamed Noureldin1, Medhat Haroun2, Iman Diab3, Sabah Elbanna4, Nashwa Abdelwahab5
1Department of Pharmacology and Toxicology, Faculty of pharmacy, Pharos University, Alexandria, Egypt
2Department of Biotechnology, Institute of Graduate Studies and Research, Alexandria University, Alexandria, Egypt
3Department of Medical Biochemistry, Faculty of Medicine, Alexandria University, Alexandria, Egypt
4Department of Environmental Studies, Institute of Graduate Studies and Research, Alexandria University, Alexandria, Egypt
5Departement of Chest Diseases, Faculty of Medicine, Alexandria University, Alexandria, Egypt
Correspondence to: Mohamed Noureldin, Department of Pharmacology and Toxicology, Faculty of pharmacy, Pharos University, Alexandria, Egypt.
| Email: |  |
Copyright © 2014 Scientific & Academic Publishing. All Rights Reserved.
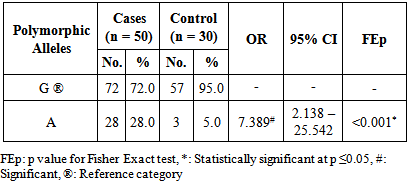
Background: Asthma is a chronic, inflammatory disease of the respiratory tract, which is characterized by bronchial hyperreactivity and respiratory obstruction in which many cells and cellular elements play a role. Interleukin 13 (IL-13) is a type 2 helper T cells (Th2) cytokine that has been shown to be pivotal in the induction of allergic inflammation of the airways. Moreover, Several IL-13 genetic polymorphisms have been linked with susceptibility to bronchial asthma. Aim: The aim of this study was to evaluate the association of IL-13 + 2044G/A polymorphism with bronchial asthma development, severity and total serum immunoglobulin E (IgE) levels in Egyptian patients with bronchial asthma. Subjects and methods: This study included 50 asthmatic patients and 30 controls. Restriction Fragment Length Polymorphism (RFLP) PCR was used for the analysis of the studied polymorphism. Also, the serum was analyzed for total IgE levels by enzyme-linked immunosorbent assay (ELISA). Results: The frequency of the A allele was significantly higher in asthmatic patients as compared to control subjects. On the contrary, the G allele was less frequent in the asthmatic patients as compared to control subjects. In addition the A allele was more frequent in severe asthmatic patients and on the other hand the G allele was less frequent in severe asthmatic patients. Moreover, the presence of the A allele was significantly associated with higher total serum IgE levels whether in homozygous or heterozygous state. Conclusions: The results showed that the IL-13 +2044 A allele is a risk factor for bronchial asthma. There is a significant association between the A allele and more severe asthma, however the G allele is significantly associated with less severe asthma. In addition, the A allele is significantly associated with higher total serum IgE levels.
Keywords: IL-13 polymorphism, Genetics, IgE, Asthma development
Cite this paper: Mohamed Noureldin, Medhat Haroun, Iman Diab, Sabah Elbanna, Nashwa Abdelwahab, Association of Interleukin 13 +2044G/A Polymorphism with Bronchial Asthma Development, Severity and Immunoglobulin E levels in an Egyptian Population, Clinical Medicine and Diagnostics, Vol. 4 No. 4, 2014, pp. 71-77. doi: 10.5923/j.cmd.20140404.03.
Article Outline
1. Introduction
- Asthma is an inflammatory disorder of the lungs that affects people of all ages and is a significant source of morbidity and mortality worldwide [1]. It is a common, chronic respiratory disease affecting 1–18% of the population in different countries. It is a heterogeneous disease, usually characterized by chronic airway inflammation. In addition, it is defined by the history of respiratory symptoms such as wheeze, shortness of breath, chest tightness and cough that vary over time and in intensity, together with variable expiratory airflow limitation. These variations are often triggered by factors such as exercise, allergen or irritant exposure, change in weather, or viral respiratory infections [2]. Although it is known that genetic susceptibility contributes to the risk of asthma, only few linkage or association genetic studies have been performed in non-European populations [3].Human Interleukin-13 (IL-13) is a 17-KDa glycoprotein cloned from activated T cells. The IL-13 gene is located on chromosome 5q31 in the cluster of genes encoding IL-3, IL-4, IL-5, IL-9 and Granulocyte macrophage-colony- stimulating factor. The gene consists of 2937 bp DNA and transcribes into 1287 bp mRNA. It has four Exons and five introns [4]. IL-13 is a Th2 cytokine that has been shown to be pivotal in the induction of allergic inflammation of the airways [5, 6]. Expression of IL-13 was upregulated at protein levels in sputum or peripheral blood of patients with asthma, and this was significantly associated with asthma-related traits, such as increased eosinophil counts and bronchial hyperresponsiveness [7-9]. The involvement of IL-13 in the pathogenesis of asthma has been extensively documented [5, 10]. Moreover, Several IL-13 genetic polymorphisms have been linked with susceptibility to develop atopic disease, such as bronchial asthma [11, 12]. In addition, many single nucleotide polymorphisms (SNPs) have been identified in the human IL-13 gene. Most of these polymorphisms are in the 5’ regulatory region or in the non-coding intron region of the gene. So far only one nonsynonymous SNP has been identified which is located in the Exon 4 of the gene at the position 2044 and changes the amino acid arginine (Arg) to glutamine (Gln) [11].The present study aimed to elucidate the association of (+2044G/A) IL-13 gene polymorphism with bronchial asthma development, severity and total serum immunoglobulin E (IgE) levels in Egyptian patients with bronchial asthma.
2. Subjects and Methods
- Ethical clearance for conducting the study on human blood samples was granted by the Ethical committee, faculty of medicine, Alexandria University, Egypt.
2.1. The Study Population
- This study included 80 subjects with age over 18 years old. The subjects were divided into two groups; group I included 50 patients with bronchial asthma (cases) and group II included age, sex and body mass index (BMI)-matched 30 normal healthy non-smoker subjects (control). The patients were selected from the outpatient clinic of both Alexandria Main University Hospital and The Maamora Chest Hospital. All patients with cardiac, renal, hepatic diseases were excluded from the study.The patients with bronchial asthma were subdivided into 18 patients with mild bronchial asthma, 19 patients with moderate bronchial asthma and 13 patients with severe bronchial asthma. Patients were selected and subdivided according to the Global Initiative for Asthma (GINA) guidelines [13].After taking an informed consent, all subjects were subjected to detailed history taking, complete clinical examination, Routine laboratory investigations and Pulmonary function tests (Pulmonary function tests were done by using CHESTGRAPH HI-701 (version 1)).
2.2. Sample Collection
- Two milliliters (mL) venous blood was collected in tubes containing EDTA as whole blood for Extraction of DNA. For serum, 3 mL of blood was taken in a separate tube, kept standing in the tube for a few minutes and centrifuged at 10,000 xg for 10 minutes, and then serum was separated. The whole blood and serum samples were kept at -20°C till further analysis.
2.3. DNA Extraction
- DNA was purified from whole blood using a spin column protocol [14] according to the manufacturer’s instructions of the GeneJETTM Genomic DNA Purification Kit.
2.4. Identification of the IL-13 +2044G/A Genotypes [15]
- Genotyping of the +2044G/A polymorphism was performed by PCR-RFLP using forward primer 5'-CTT CCG TGA GGA CTG AAT GAG ACG GTC-3' and reverse primer 5'-GCA AAT AAT GAT GCT TTC GAA GTT TCA GTG GA-3' [15]. Dream TaqTM Green PCR Master Mix (2x) was used for amplification. (Fermentas, Canada). The master mix contained Dream TaqTM DNA polymerase in 2x Dream TaqTM green buffer, deoxynucleoside triphosphates (dNTPs), and 4 mM magnesium chloride.PCR was carried out in a thermal cycler in a total volume of 50 μl containing 25 μl of 2x Master Mix, 1 μl of each primer (50 picomoles), 5 μl of the extracted DNA (20 ng), 18 μl of nuclease-free water. The PCR conditions were as follows: initial denaturation step of 5 minutes at 95°C, 35 cycles of 30 seconds at 95°C for denaturation, 1 minute at 57°C for annealing, 40 seconds at 72°C for extension, and a final extension step of 10 minutes at 72°C.The products of PCR were cleaved with NIa IV as follows: 25 μl of the PCR product were added to 1 μl of the enzyme and then 4 μl buffer and 10 μl sterile distilled water were added, followed by incubation at 37°C for 1 hour and then electrophoresed on 2% agarose stained with ethedium bromide. This digestion produced 178 and 32 bp fragments for +2044G allele, while produced a single 210 bp band for the +2044A allele. A 302 nm ultraviolet transilluminator (Biometra, Germany) was used for visualization of the DNA bands and they were photographed.
2.5. Immunological Investigation [16]
- The serum was analyzed for total IgE levels by enzyme- linked immunosorbent assay (ELISA) using the commercially available kit (Calbiotech IgE ELISA kit) according to the manufacturer’s instructions.
2.6. Statistical Analysis
- Data were analysed using IBM SPSS software package version 20.0. Qualitative data were described using number and percent. Comparison between different groups regarding categorical variables was tested using Chi-square test. When more than 20% of the cells have expected count less than 5, correction for chi-square was conducted using Firsher’s Exact test or Monte Carlo correction.For abnormally distributed data, Kruskal Wallis test was used to compare between different groups and pair wise comparison was assessed using Mann-Whitney test taking into consideration Bonferroni correction. Odd ratio (OR) and 95% Confidence Interval were used to calculate the ratio of the odds of an event occurring in one patient group to the odds of it occurring in the control group.Significance test results are quoted as two-tailed probabilities. Significance of the obtained results was judged at the 5% level.
3. Results
3.1. Demographic Data
- No significant differences were observed between the two studied groups as regards sex, age and BMI distribution.
3.2. Serology
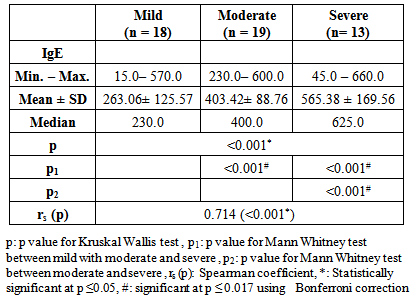
- In the present study the asthmatic patients had significantly higher IgE levels than the controls (P<0.001) (figure 1). In addition a positive correlation between IgE levels and severity of asthma was observed (table 1).
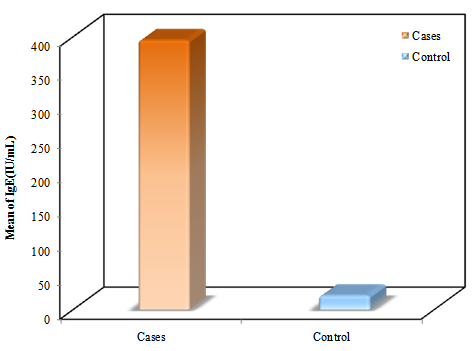 | Figure 1. Comparison between the two studied groups according to IgE levels |
|
3.3. Molecular Findings
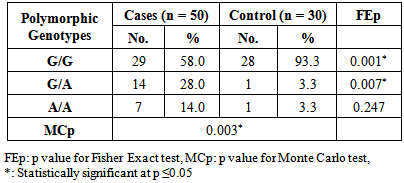
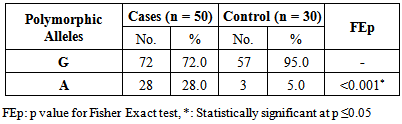
- As shown in figure 2, lane 6 represents G/G genotype with one band at 178 bp. lanes 1 and 8 represent A/A genotype with a single band at 210 bp. lanes 5 and 7 represents G/A genotype with two bands at 210 bp and 178 bp. lane 3 represents a negative control, while lane 4 represents Gene rulerTM Ultra Low Range DNA Ladder, fermentas, Canada. The 32 bp fragment was too small to be resolved on the agarose gel.
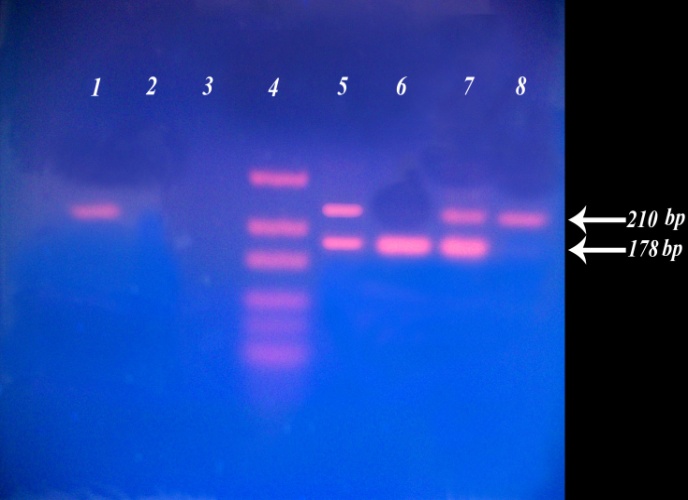 | Figure 2. The IL-13 +2044G/A polymorphism on agarose (2%) gel after digestion by NIa IV enzyme |
|
|
|
|
|
|
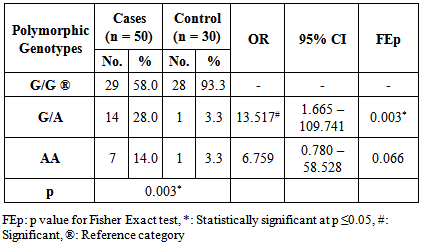
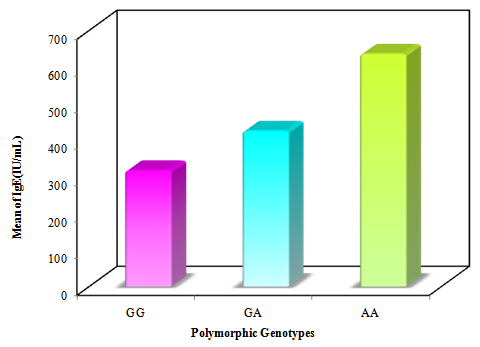 | Figure 3. Comparing IL-13 +2044G/A polymorphic genotypes in relation to total IgE levels in asthmatic patients |
4. Discussion
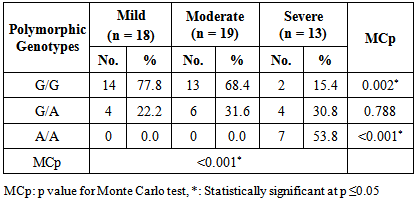
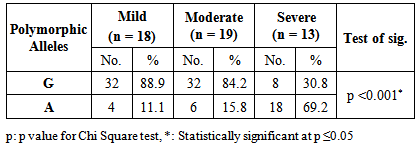
- Asthma is a chronic inflammatory disorder of the airways in which many cells and cellular elements play a role [17]. Polymorphisms that may dysregulate the function and/or the expression of IL-13 occur frequently in the population and have been consistently associated with allergic phenotype [18]. Arima and colleagues have showed an evidence that the IL-13 +2044G/A may be a functional variant. Utilizing a mutant recombinant IL-13, they showed that recombinant IL-13 containing the G1n 110 variant (resulting from A allele) bound the IL-13 Rα2 chain with a lower affinity than the wild type IL-13, resulting in a lower clearance rate of the cytokine. In addition the asthmatic patients homozygous for the Gln 110 variant had higher serum levels of IL-13 than those without the variant. They postulated that impaired binding of IL-13 by the soluble IL-13 Rα2 chain leads to a higher serum levels and prolonged activity of IL-13 in vivo [19].The present study showed a strong significant association between IL-13+2044G/A polymorphism and asthma risk regarding the A allele in an Egyptian population.A meta-analysis was conducted in the Chinese population, showed that IL-13+2044G/A polymorphism was associated with asthma risk in Chinese adults [20]. Furthermore, another meta–analysis based on thirty studies with 12,781 asthmatics and 11,500 controls, showed an association between IL-13+2044G/A and susceptibility to asthma (p = 0.04) [21].Our results come in accordance with a case-control study on Egyptian children where asthmatic children had significantly higher frequency of the heterozygote genotype GA (50%) than controls (15%) fisher's test = 0.0407 [22]. Moreover, Heinzman et al. conducted case control studies in a British population (150 young adults with asthma and atopy and 150 controls) and a Japanese population (100 young adult with asthma and atopy, 100 subjects with non-atopic asthma and 100 controls), in the British population the A allele was significantly associated with asthma (P = 0.014), while it was significantly associated with both atopic asthma (p = 0.033) and non-atopic asthma (p = 0.047) in the Japanese population [23].Recently, Yang et al. [24] reported that the A allele of IL-13 +2044G/A showed a 1.38-fold significantly increased risk for development of asthma (P = 0.040) in an adult Chinese population. Furthermore, Bottema et al. [25] conducted their study on three Dutch Caucasian populations (Rhinitis trios, Asthma trios and asthma case control population) and in line with our results they found an association between lL-13 + 2044G/A polymorphism and asthma in both asthma populations. Where, in the asthma case-control population, genotypes with 1 or 2 copies of the A allele were significantly more prevalent in asthmatics than in controls (p = 0.005). [25] Moreover, the A allele has been reported to have a great relationship with allergic asthma in Iranian population [26]. In addition, WU et al. [27] carried out a pediatric study on asthma in middle china, they showed that the A allele of IL-13+2044G/A was significantly associated with increased risk of asthma (P = 0.0010). Opposite way round, Wang et al. [28] did not to find any significant association between IL-13+2044G/A polymorphism and asthma in Taiwanese population. In addition Chan and associates [29] in a pediatric study on Chinese asthmatics showed no significant difference in genotype and allele frequency between the cases and the control groups. Also, a similar result was shown in Dutch [12] and Korean [30] populations.Absence of association between IL-13 +2044G/A and asthma was reported by Lopez et al. [31] in the Mexican population. Furthermore, Celedon and associates [32] found no significant evidence of linkage disequilibrium between IL-13 +2044G/A and asthma in a family – based association study in Costa Rica.A study conducted in Korean children population did not find any evidence that IL-13 + 2044 G/A polymorphism was associated with asthma susceptibility or atopy [33]. Furthermore, Dimitreva – Zdorova et al. [34] showed no association between IL-13+2044G/A polymorphism and atopic bronchial asthma development in a Russian population.These different results of the above studies may be due to the study design and criteria of the enrolled subjects such as circumstantial environment. Furthermore, gene-to-gene and gene-to-environment interaction may have a role in these results. Also, environmental factors may exert significant influences on the pathogenesis of asthma in adults. As some environmental factors may imitate some mutation genes, which result in asthma symptoms in adults. So it is more difficult to confirm the effects of genetic factors on asthma in adults.Regarding asthma severity, in the current study we observed a significant strong association between the A allele of IL-13+2044 G/A polymorphism and more severe asthma (p< 0.001), while G allele was significantly more prevalent in less severe asthmatics (mild patients) (P< 0.001).In consistent with our results, Beghe et al. [35] reported borderline association (P = 0.006) between IL-13+2044G/A polymorphism and asthma severity including the risk allele (+2044A). On the other hand, other studies revealed no association between IL-13+2044G/A and asthma severity in Egyptian children [22], Chinese children [36] and Russian [34] populations.IgE molecules have been proven to play a crucial role in allergic respiratory diseases and may lead to chronic airway inflammation in asthma through activation of effector cells via high affinity FcεRI or low affinity FcεRII IgE receptors [37].In the current study, the patients harboring one or more copy of the A allele were found to have significantly higher IgE levels compared to those carrying the wild genotype (GG). Thus it can be suggested that there is a strong association between the A allele and serum elevated IgE levels. In consistent with our results, Graves et al. [11] reported that the A allele is strongly associated (p = 0.000002) with increased serum IgE levels in 3 different populations comprising a total of 1399 children. In addition the A allele was reported to be significantly associated with increased serum total IgE in a Costa Rican population (p < 0.05) [38]. Furthermore, El-sayed et al. [22] showed that serum total IgE was significantly higher in asthmatics with GA genotype as compared to those with GG genotype in asthmatic Egyptian children. In addition Mijin kang et al. [39] in a pediatric study reported significantly higher total IgE levels in asthmatic patients carrying one or two copies of the IL-13+2044A versus those homozygous for +2044G (P = 0.011).On the other hand, no association between this polymorphism and IgE levels was reported in Korean [33] and Russian [34] populations.
5. Conclusions
- There is a positive correlation between total serum IgE levels and asthma severity. The presence of IL-13 +2044 A allele is a risk factor for bronchial asthma. Moreover, there is a significant association between the A allele of IL-13 +2044G/A polymorphism and more severe asthma. In addition, IL-13 +2044 A allele is significantly associated with higher total serum IgE levels.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML