-
Paper Information
- Next Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Clinical Medicine and Diagnostics
p-ISSN: 2163-1433 e-ISSN: 2163-1441
2012; 2(5): 45-50
doi: 10.5923/j.cmd.20120205.01
Numerical Chromosomal Abnormalities in Patients with Acute Lymphoblastic and Myeloid Leukemia in Iran
Ali mohamad malekasgar 1, Mohamad Pedram 2, Sayyed kamal Eshagh housaini 3
1Dept. of basic science, genetic unit, qom university of medical sciences, Iran
2thalassmia and hemoglobinophaties research center, Ahvaz-Iran Ahwaz Jondishapur University of Medical Sciences, Iran
3Dept. of oncology, Qom university of medical sciences, Iran
Correspondence to: Ali mohamad malekasgar , Dept. of basic science, genetic unit, qom university of medical sciences, Iran.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
The majority of cases of ALL demonstrate an abnormal karyotype, either in chromosome number or structural changes. Abnormal chromosome number in childhood acute lymphoblastic leukemia defines distinct biological subgroups with a different response to treatment. The tubes are cultured with three different protocols to save time if one protocol failed. Cultures are then harvested, and cells are fixed and chromosome spreads are prepared. Of 25 patient studied, one patient had psudodiploid karyotype, three patients had tetra-ploid karyotype, four patients had low hypo-diploid karyotype, four patients had high hyper-diploid karyotype, five patients had low hyper-diploid karyotype and eight patients had normal karyotype. Some other factors like, Age, Sex, Consanguinity, Hemoglobin, WBC count, and Type of the leukemia cell also have been evaluated. We found excess number of patients having hypodiploid karyotype but still response to treatment protocols were satisfactory. By comparison between 4 different cultures methods, we find direct method to be more efficient for ploidy analysis.
Keywords: ALL, AML, Karyotype, Ploidy, Leukemia
Cite this paper: Ali mohamad malekasgar , Mohamad Pedram , Sayyed kamal Eshagh housaini , "Numerical Chromosomal Abnormalities in Patients with Acute Lymphoblastic and Myeloid Leukemia in Iran", Clinical Medicine and Diagnostics, Vol. 2 No. 5, 2012, pp. 45-50. doi: 10.5923/j.cmd.20120205.01.
Article Outline
1. Introduction
- Acute lymphoblastic leukemia (ALL) is characterized by clonal proliferation, accumulation, and tissue infiltration of neoplastic cells. They are mainly regarded as childhood diseases, with an early incidence peak at two to five years of age, where they represent about 80% of the childhood leukemia in the United States, and occur with an incidence of 30 cases per one million population per year[1]. A basic principle of the WHO classification is that the diagnosis of myeloid and lymphoid malignancies should be based on the integration of the information derived from genetic, immunophenotypic, biological, and clinical features in order to better define specific disease entities. In fact, genetic findings may predict the prognosis and biologic properties of the leukemia more consistently than does morphology[2]. Chromosomal abnormalities in childhood ALL had important significance related to prognosis, diagnosis and management. Chromosomal classification in ALL is based on the number of chromosomes in abnormal cells (ploidy pattern) as well as the structural changes.Abnormal chromosome numbers in childhood acute lymphoblastic leukemia, defines distinct biological subgroups with a different response to treatment[3,4]. The largest subgroup within the non-T cells ALL (approximately 25%) is the hyper-diploid group, which is associated with favorable characteristics and outcome[3,4,5]. In contrast the hypo-diploid and near-haploid ALL is much rarer (1%) and has shown a very poor outcome with the few intensive protocols reported[6,7]. Near haploid has been reported in around 41 children[5,8,9] and in six additional children within a clinical study[9]. The majority of cases of ALL demonstrate an abnormal karyotype either in chromosome number or as structural changes such as trans-locations, inversions, deletions, etc. These changes were detected in only half of ALL patients in the first banding studies[10]. The scantiness of information gained from chromosomal findings in ALL has been in large part, due to technical difficulties. The emerging theory for the role of constitutional trisomy 21 in leukaemia predisposition is that genes on this chromosome contribute to the expansion of haematopoetic compartments during early development that result in an increased pool of potential tumour precursor cells[11].Chromosome studies in ALL exhibit poor morphology. Chromosomes tend to spread poorly, and appear blurred and fuzzy with indistinct margins, making banding studies challenging or even impossible[12,13] Improvements in spreading and banding techniques have resulted in higher rates of detection, and most studies now report chromosomal changes in 60% to 85% of ALL cases.[14,15]. We have conducted this study to evaluate numerical abnormalities in these patients for better management of patients admitted to this hospital by using different cell culture and chemotherapy treatment protocols.
2. Material and Methods
- This work was conducted on 25 cases (five cases with AML and 19 cases with ALL) referred to our laboratory for karyotype and ploidy determination of chromosomes after getting a written consent from their parents. Only new diagnosed cases (from December 2007 to July 2008) were included in this survey and old cases which have been already started chemotherapy treatment were excluded. 0.5-1 ml of bone marrow aspirate were collected in centrifuges tube containing five ml RPMI complete with 1% preservative-free heparin. In few cases where the bone marrow samples were unavailable or failed to grow in culture, blood samples were used. The optimum cell density for a bone marrow culture is 106 cell/ml. All samples were optimized for cell density, using hemocytometer (slide counter chamber). In most cases, the preferred culture method was overnight colcemid (ONC), followed by flurodeoxy uridine (FudR) and 24 hour protocols. In AML samples the preferred technique was FUdR, followed by 24 hours cultures and then ONC. For each patient we set up at least three culture tubes including one direct cultures method[12] and two out of the three fore-mentioned methods. Quality of spreads in these methods were compared. Good and analysable spreads could obtain with direct method for ploidy determination in most cases, but for structural chromosomal analysis, flurodeoxy uridine (FudR) method proved to be satisfactory.
3. Results
3.1. Number of Patient, Age, Sex, Ethnic Groups, Consanguinity, Hemoglobin Rate and WBC Count Factors
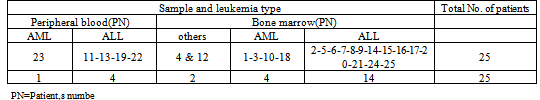
- Bone marrow (19 patient) or blood (six patients) samples from 25 patients, suffering from leukemia referred from Shafa hospital have been analyzed. Among 25 patients participated, 19 ALL (including two cases with T-ALL and seven cases with B-ALL), five AML and one unidentified case was present.Out of 25 patients, 16 males (64%) and nine females (36%) are recorded. The age of patients ranges from 1.5 to 12 with median age of 5.82 years. Of 25 parents participated, eight couples (32%) have not reported the type of their marriage, (consanguinity) and out of remaining 17 (68%) couple, 11 couples (44%) had consanguineous marriage (10 firsts and one second) and six couples (24%) had non-consanguineous marriage. Table 1
|
3.2. Response to Treatment
- Of these 25 leukemic cases, 23 (92%), achieved complete remission but with different resistant. Two patients (8%) did not respond to treatment and survival, one of them with hypo-diploid karyotype had Philadelphia chromosome, and other one had complex karyotype of hypo-diploid and near-haploid . Both of them were among AML patients with M3 and M4 subgroup, Table 3. Regarding hematological parameters, cases with lower Hb (< 8 gm/dl) and high WBC (>50000/mm3) showed a better remission rate. Cases with age < 5 years, male sex and couples with positive consanguinity also showed a better remission rate, although not significant.
3.3. Clonal Abnormalities
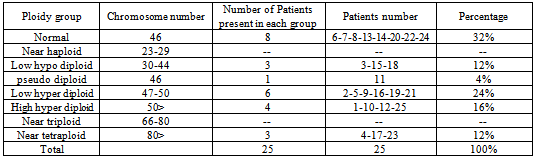
- Of 25 patients studied, 17 (68%) cases with clonal ploidy abnormalities and eight cases (32%) of normal karyotype were recorded. Among 17 cases with more than one abnormal karyptype, 10 cases (40%) were hyper-diploid,[six low (47-50 chromosomes) and four highs (> 50 chromosomes) hyper-diploid], three cases (12%) were hypo-diploid (30-44 chromosomes), Three cases (12%) were near tetra-ploid, (> 80 chromosomes) Figures 1,2 and 3. and one patient was pseudo-diploid, Table 2.Although some of these patients are hypo-diploid (three cases), or pseudo-diploid (one case), most of them (92%) had good remission with treatment protocols (U.K.C.C.S.G) for ALL, and (B.F.M) AML patients. Only two patients (8%) had no responses to treatment, both of which were among AML leukemic cases.Of 17 patients with abnormal karyotype, eleven B-cells ALL and five AML with M1 to M5 subgroups were recorded. Classification of one of the patient could not be determined. Eight patients had complex karyotype with more than two clonal chromosomal abnormalities. Details are available on request.
|
|
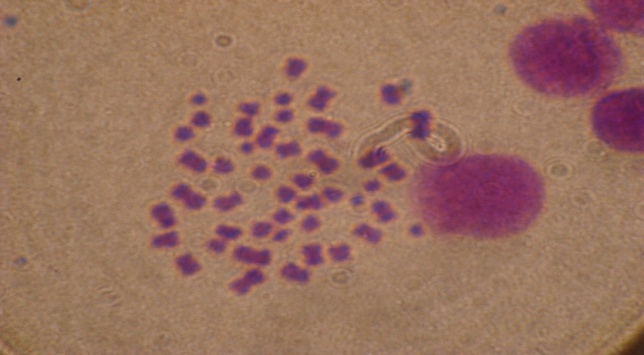 | Figure 1. Patient No. 5 with low hyperdiploidy(48 chromosome) |
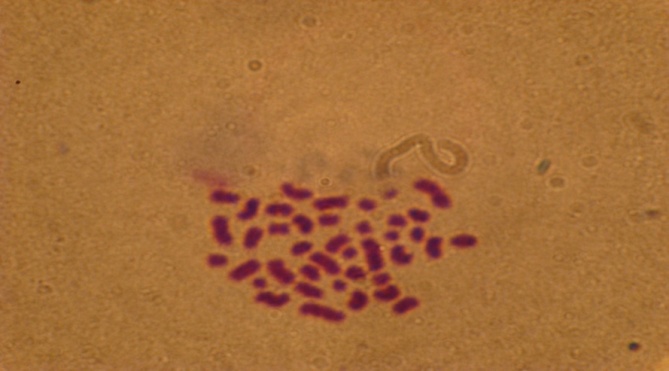 | Figure 2. Patient No. 15 with low hypodiploidy (40 chromosom) |
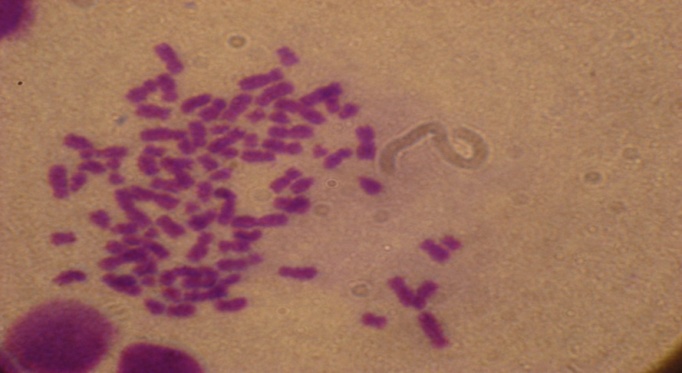 | Figure 3. Pationt No, 23 with near tetraploidy(83 chromosome) |
4. Discussion
- Most of the leukemia patients have cytogenetical or molecular abnormality and it is shown that there are some relations between chromosomal abnormalities and morphological and immunological characteristics of the cancer cells in these patients. More important are karyotype changes which show important prognosis without any relations to other variables like age, sex and primary leukocyte count, and so permits to differentiate between high risk and low risk patients at the time of diagnosis for proper treatment.Cytogenetic analysis of 25 cases in our study shows 8 patients (32%) to have normal diploid karyotype, similar to many reports . Pseudo-diploid on the other hand were found in only one case (4%) which is much lower than the earlier reports which suggested that pseudo-diploid comprised the largest cytogenetic group (41.5%) in childhood ALL[16]. This could be due to misclassification of pseudo-diploid karyotype by failure of detection of some submicroscopic genetic abnormalities at the conventional cytogenetic analysis. Low hypo-diploid karyotype (30-44 chromosomes) were present in 12% of the our cases, which are higher than those reported by others who considered hypo-diploidy to be a relatively uncommon (>9%) finding in ALL[17,18]. Many studies reported low percent of patients in hypo-diploid group, but some studies[19-20], including our study (12%) found much higher percent of the patient in this group with good remission. In studies in which only cytogenetic investigations are carried out, some hypo-diploid cells may not diagnosed correctly due to presence of many hyper-diploid cells and only after relapse of the disease and repeat investigation of these patients with FISH, or other advanced molecular methods, the true karyotype can be detected. Hyper-diploid (low + high hyper-diploid) represented a majority of ALL cases in our study and many other authors have also reported same higher frequency[21,22].The remission rates in our study cases were almost same among hypo-diploid, pseudo-diploid and hyper-diploid groups, but the normal diploid cases had better remission rate. Diploid group was reported to have the best remission by some authors[19] although others considered it to be of an intermediate remission rate[23,24]. Response to treatment rates varied among chromosome ploidy groups. The highest response rates were seen in patients with normal karyotypes and a modal number >50 chromosomes. In our study, after normal karyotype cases, we also found the best remission rate in this group. Hyper-diploidy was found by some authors to have a good prognosis[25,26] unless associated with structural aberrations making it less favorable[19,27,28]. In our study, we had two resistant cases, one mixed numerical aberration (near haploid + low hypo-diploid) and the other one with low Hypo diploid, both of which were among AML subtype. Near-haploid (< 30 chromosomes) ALL, were not present in our case, except the one with mixed numerical aberration (near haploid + low hypo-diploid) between two resistant patients. Near-haploid is a rare and unique subgroup associated with a very poor outcome. It may be underestimated being masked by a coexisting hyper-diploid line. In our study although any chromosome could be involved in tetrasomy, most of chromosomes especially 5,9,10, 12,18, 20,22 and X were duplicated more frequently.Of four Majors Ethnic groups in Khozestan province including Persians, Iranian Arabs, Bakhtiari Lurs, and Behbahanis, 14 Arab (56%), 9 Lurs (36%) and two Farsis (8%) are recorded. Prevalence of Arabs were significant in these four major ethnic groups, although it may be unreliable due to small sample size, Table 1. We could not find any data showing prevalence of leukemia in these Ethnic groups in Iran. The most common FAB type among our cases (48%) were L2 subtype with better remission compared to other immunophenotypes[29]. Some studies reported a poor prognosis associated with L2 and other studies reported that significance of L2 remained controversial[30]. Although full analyzing of karyotype spreads for determination of structural, numerical and submicroscopic genetic abnormalities at the conventional cytogenetic analysis and its easier detection by the molecular and FISH technique is mandatory, but determination of only numerical chromosomal changes (ploidy pattern) for typing of patients into different ploidy groups can also be useful as a primary step. Therefore we decided to conduct this study because of increasing number of ALL patients and need for immediate report for ploidy determination. We compare three standard methods including, overnight colcemid (ONC), flurodeoxy uridine (FudR) and 24 hour protocols, with a modified direct culture method (12), and found the modified direct culture method to be easy and satisfactory for ploidy pattern determination in most (68%) cases.
5. Conclusions
- We found excess number of patients having hypo-diploid karyotype but still response to treatment protocols were satisfactory. Variation in number of hypo-diploidy in different studies could be due to difference in ethnic group. Out of four different cultures methods used, we found the direct method to be satisfactory for ploidy analysis.
ACKNOWLEDGEMENTS
- The authors of this article appreciate the authorities of Ahvaz Jundishapour university of medical science for their financial support of this work. We also wish to thanks Dr. Mohammad Housain Sarmast, the dean of Ahvaz Joundishaour University of Medical Science for his assistance and support. Special thanks to Dr. ALI Gasemi and Mrs. Nafiseh Esfandyari and all the members of children Oncology unit of Shafa Houspital for their valuable contribution.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-Text HTML
Full-Text HTML