-
Paper Information
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Clinical Medicine and Diagnostics
p-ISSN: 2163-1433 e-ISSN: 2163-1441
2012; 2(4): 37-44
doi: 10.5923/j.cmd.20120204.04
Clinical Genomic Analysis and Diagnosis --Genomic Analysis Ex Vivo, in Vitro and in Silico
Biaoru Li
Artemis Health, Stanford University, Menlo Park, CA 94025 and Dept. of Pediatrics, MCG, GHSU, 1120 15th St. CN-4111, 30912, Augusta, GA
Correspondence to: Biaoru Li , Artemis Health, Stanford University, Menlo Park, CA 94025 and Dept. of Pediatrics, MCG, GHSU, 1120 15th St. CN-4111, 30912, Augusta, GA.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
Seven years ago, I systemically reviewed single cell techniques with genomic and proteomic analyses which was called Single-Cell Genomic Analysis. After many years of arduous work, single cell techniques with downstream genomic and proteomic analysis have been applied to clinical fields including molecular pathology, molecular genetics, forensic medicine and biomarker discovery. On top of that, dynamic cell-sorting technique combined with downstream cell culture and genomic analysis of stem cell for regeneration medicine and cancer stem cell for differentiation have also been greatly developed in clinical fields. More importantly, tissue level sampling with in silico analysis has been applied in therapeutic targeting for advanced neoplastic disease. Recent development in sorting homogeneous cells in vitro (or single cells technique), ex vivo (dynamic analysis or small number of cell culture with downstream genomic analysis) and insilico (tissue level sampling with in silico analysis) have allowed physician scientists with a choice to select one of these above techniques with genomic analysis to apply to their clinical research fields. To fully understand these modern techniques, this manual will review recently developed methods or clinical genomic analysis in vitro, in silico and ex vivo. In the review paper, I will also introduce how to utilize these techniques in different clinical fields. The manual will also address some of the challenges for clinical genomics analysis and diagnosis due to mixed cells from clinical specimens.
Keywords: Genomics Analysis, Clinical Genomics Diagnosis, Single Cell Diagnosis, Single Cell Genomics Analysis , Diagnosis, Primary Cell Culture, Biomarker Discovery, Therapeutic Targeting
Article Outline
1. Introduction
- The mixed cell population in clinical samples can mask the actual results of genetic diagnosis and genomics analysis. In order to overcome the challenges in purity and limited cell counts after initial purification, single cell technique, or similar techniques, have now being widely developed for over a decade. One of the most common is single cell PCR[1]. Due to advancements in techniques in amplifying genomic DNA/RNA and signal magnifying for protein from small number of cells, these techniques, including genomics and proteomics are now widely being employed in the clinical fields[2]. Based on research and development for genomic analysis from clinical samples, physicians and scientists are studying faster/purer techniques, such as laser capture microscopy, along with downstream genomic analysis to study clinical specimens. The single-cell harvest obtained from glass slides combined with downstream genomics and proteomics have been developed in molecular pathology, molecular genetics and forensic medicine[3]. Some call thetechnique, cytogenomics[4]. The technique is named as in vitro analysis because the procedure defined as in vitro (Latin: within the glass or glass slide) is performed outside the living organism. On the other hand, dynamically pure cell-sorting technique combined with ex vivoculture with downstream genomic analysis (such as stem cell for regeneration medicine and cancer stem cell for inducing differentiation) has also been tremendously developed[5]. This technique is known as ex vivo because the measurements are done in an artificial growth environment (outside the organism) with minimum alterations of the natural conditions. Moreover, after Dr. Schmid first used “micro-dissection in silico” to analyze gene expression profiles to uncover biomarkers from clinical specimens[6], tissue level sampling by in silico analysis is now increasingly being applied in genomic analysis of heterogeneous cells from clinical tissues[7]. Here the technique is called as in silico genomic analysis due to the performance via computer or program simulation, as an analogy to the Latin phrases in vivo and in vitro refer to experiments done in living organisms and outside of living organisms, respectively. In order to incorporate all the clinical genomic techniques by using these three methods, in vitro, ex vivo and in silico, I will refer to clinical genomic analysis as genomic analysis and diagnosis by in vitro, ex vivo and in silico. Genomic analysis in vitro, used in molecular pathology and molecular genetics from pretreated tissue[8], combines cell isolation and genomic analysis from clinical specimen. Genomic analysis ex vivo, encompass three processes, 1) living cell separation, 2) culture and 3) downstream genomics analysis as well. Now, FACS/MACS for living cell separation can be combined with fixed cell immunohistochemical staining (IHC)[9] and laser micro-dissection technique (originally only used for fixed cell) can be utilized in living cell with downstream cell culture[10]. Genomic analysis in silico, direct tissue-level specimens for genomic analysis, in which several modules in bioinformatics are able to identify specific genomic profile in the mixed tissue level, Here, in order to better understand clinical genomic analysis and diagnosis from clinical specimens, I will compare genomic analysis among intro, ex vivo and in silico for different clinical specimensand different purposes. Based on their conditions and purposes, clinical scientists can decide which way is the best genomic analysis for their specimens.
2. Clinical Genomic Analysis by intro, ex vivo, and in Silico
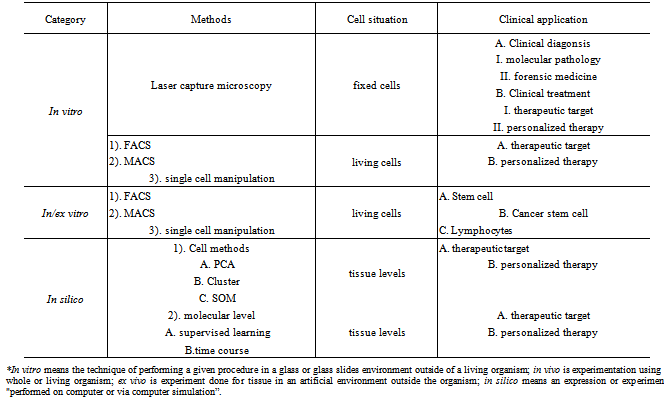
- In my previous review paper[11], it was shown that the flow-cytometric cell sorting, magnetic cell separation (FACS/MACS) and laser-based micro-dissection of tissues provide the basic methods to isolate similar cells to study gene expression profiling from clinical specimens. FACS isolating technique for cells in solution labelled with fluorescent signals can sort these cells with a specific biomarker such as CD3/CD4/CD8 for lymphocytes and CD133/CD34 for stem cells and cancer stem cells (CSCs). At present, multi-colored fluorescence-activated cell sorters (multi-coloured FACS) can selectively separate and collect homogeneous cells with identical phenotypic features in a collection tube so that FACS can increase its ability to study gene expression profiling in a given cell type[12]. In the other fields, laser micro-dissection technique to isolate cells on glass slides labelled with fluorescent signals or markers can sort clinical cells that rely on specific mRNA/protein biomarkers and morphology change such as tumor cells or cancer stem cells. The characteristics of laser micro-dissection have allowed us to quickly study a given cell in vivo localization and to analyse the cell’s microenvironment in vivo[13]. Relied on research and development, the techniques for sorting single-cell or a small number of homogeneous cells from clinical specimens have been developed and categorized by the methods, purposes and functions described in Table 1. Here I will introduce each method,in details, along with this advantage and disadvantage in the following order: (1) genomic analysis ex vivo covering cell-sorting with downstream culture ex vivo and genomic analysis; (2) genomic analysis in vitro including treated cell (fixed and stained cells) and un-treated cell and (3) direct genomic analysis in silico for clinical specimens.
|
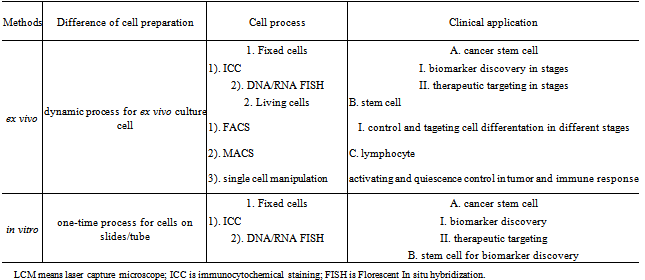
2.1. Genomic Analysisex vivo
- Cell culture from clinical specimens is different from cell lines. Cells that are cultured directly from clinical specimens are known as primary cells. The primary cells isolated from tissues can be cultured ex vivo or in vivo environments. The primary cell cultures can be often used in lymphocytes, cancer stem cell, stem cells and primary cancer cells[14]. Along with the research of stem cells for regenerative medicine, the study of cancer stem cells for biomarker discovery and the application of T-cell for passive and active immunotherapy as shown in Table 2, stem cells, cancer stem cells, and T-cells harvested from clinical specimens with genomic analysis can play an increasingly important role in biological and medical fields. Although ex vivo differentiation of stem cells intospecial somatic cell types such as neurons and myocytes has a well-established model, long-term cultureof ex vivo autogenously adultstem cells so far have not been completely successful. The extremely low number of these cells in primary hematopoietic organs and the lack of good culture systems that support proliferation of undifferentiated stem cell have influenced their biological research and clinical application. In 1999, we reported cancer cell cultured in high-dose radiated mice to increase the cell number to study the cell characteristics[15]. Encouragingly, now repopulating capacityof ex vivo culture of hematopoietic stem cells has also largely been reported, which will indicate a good future for regeneration medicine[16]. In addition, ex vivo culture and long-term storage of induced pluripotentstem cells(IPS cells) have been tremendously reported.
|
|
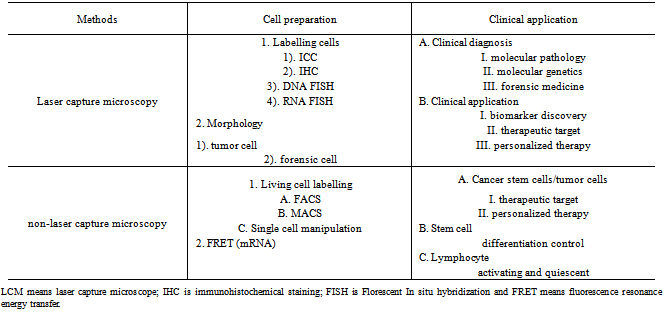
2.2. Genomic Analysis in vitro
- Since 1976, laser micro-dissection has been increasingly applied in different fields as shown in Table 3. Currently, three microdissection methods have been routinely employed. Those are (1) laser-assisted mechanical tissue micro-dissection[17], (2) laser pressure catapultmicro-dissection[18] and (3) laser capture micro-dissection[19]. Most laser-based microdissections require tissue pretreatment such as fixation, dehydration and staining. The application of these techniques requires fully consideration to downstream work such as DNA array CGH, mRNA microarray and proteomics. The advantage of laser-based microdissections can be combined with Ab-basedimmunohistochemical/immunocytochemical (IHC/ICC) or DNA FISH and RNA FISH staining to increase the cell specificity from their biomarker staining. In these several years, following developmental requirement of cancer stem cells for biomarker identification and therapeutic targeting and the application of stem cells for regeneration medicine, laser-based micro-dissection will be faced with two large developments: (1) as indicated above, process from cultured living cells, which can continue culture for downstream genomic analysis; (2) combined with automation system, whose process can be used for high-throughput screening such as for biomarker discovery[20].Cell-sorting technique by FACS and MACS is also quickly developed. With FACS technique development of multi-colored fluorescence-activated cell sorters and increasing antibody products, the FACS technique can play a much more important role in cell-sorting technique for genomics and proteomics analysis of clinical specimens[21]. Nowadays, two fields are being quickly developed, the one is based on labelling mRNA inside cells to sort cells by florescent-labelled mRNA and the other one is combined with automation system, the purpose is high throughput screening to discover new biomarkers[22].Due to impurities of FACS and MACS in clinical specimens, we have added a single cell manipulation to increase the purity after MACS or FACS process so downstream-cells can provide more accurate data in genomic signature analysis and diagnosis for patient’s therapeutic targeting[23].
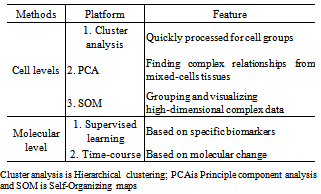
2.3. Genomic Analysis in silico
- In the clinical field, clinical specimens are often and directly frozen due to requirements of pathological processes. If the specimens are processed at the tissue level for microarray by total mRNA, array CGH by total genome DNA and proteomics by total protein, genomic analysis in silico is an exclusive way for the analysis of genomic data because the genomic data at the tissue level are mixed with genome from different cells[24]. According to our experiences, at least three ways can be used for genomic analysis for heterogeneous cells: hierarchical cluster, principle component analysis (PCA), and self-organizing map (SOM). Hierarchical clustering can be quickly processed due to its usage of similar expression patterns for cell groups. Principle component analysis (PCA) is primarily aimed at finding complex relationships between variables in a dataset so it has been extensively applied in biomarker discovery of known cells from mixed-cells tissues. This feature helps us to study variables and factors that are not correlated to each other[25]. The self-organizing map (SOM) is a powerful tool for grouping and visualizing high-dimensional complex data which can be applied for a two-dimensional plane[26]. Following extracting data from mixed cells of clinical specimens, we have routinely employed two of the three platforms to analyse data from the mixed cells as shown in Table 4. If genomic data from the clinical specimens do not contain a series of control samples to process micro-dissection in silico, we can use clinical bioinformatic module combined with biological or medical biomarkers corresponding this disease to analyse the data, such as using “supervised leaning” protocol.
|
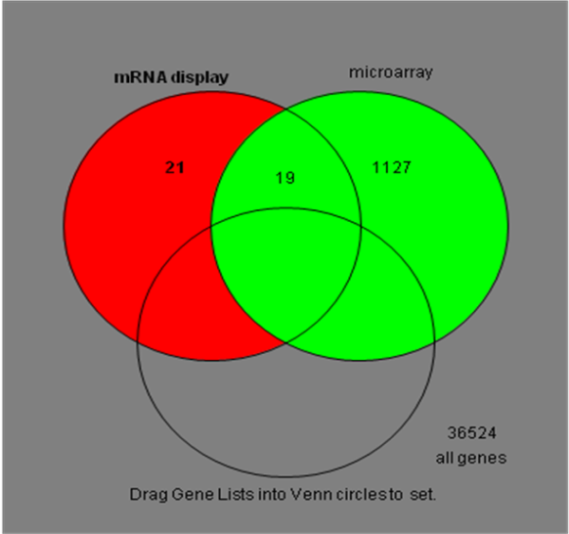 | Figure 1. Relationship between genomic analysis in vitro (single-cell genomics or single-cell mRNA display) and genomic analysis in silico (microarray in tissue level) |
3. Application of Clinical Genomic Analysis
3.1. Regeneration Medicine
- A major issue in regenerative medicine is the cell sources used to rebuild damaged tissues. Although many questions are still unanswered, most physicians and scientists nowadays have reported that regeneration in humans from the body's own tissues is feasible. Regeneration is a regulative developmental process ubiquitous across all human organs. It functions throughout the life cycle to restore the normal function of cells, tissues, organs, appendages and whole organisms.The regenerative capability is absent or low in some cells such as hepatocytes, myofibers, osteocytes, and most neurons[27]. We know that three sources of cells, induced pluripotentstem cells(IPS cells), embryonic stem cells (ESCs) and adult stem cells (ASCs) can be used for cells and/or organs regeneration for damaged tissues. ESCs are in vitro cultivated pluripotent cells derived from the inner cell mass (ICM) of the embryonic blastocyst. ES cells can be differentiated into representative derivatives of all three embryonic germ layers (endoderm, ectoderm and mesoderm) in both in vitro and in vivo[28].Adult stem cells also can multiply to regenerate the definitive cells to replace damaged tissues. They have been found in normal scaring after injury, such as in myocardium, pancreatic islet cells, spinal cord and retina tissues[29]. Because stem cells (ESC and ASC) have high growth and self-renewal capacity, several papers and protocols have been successfully reported to use the cells into the therapeutic possibilities, such as diabetes, mycardiac infarction and Parkinson’s disease. Genomic analysis ex vivo can play a critical role in proliferation and differentiation of stem cells for regenerative medicine, such as dynamic monitoring genomic profile for a small amount of cultured cells. After we understand the genome and the corresponding pathway from the primary cells in a differentiating period, we can select corresponding growth factor or compound to control cell proliferation and to target differentiation based on the genomic analysis.
3.2. Clinical Diagnosis
3.2.1. Single Cell Diagnosis Via Single Cell Genomic Analysis/Diagnosis
- The pathology diagnosis relies on cell morphology and cell arrangement such as tumor cell diagnosis based on the cell morphological change and infiltrating into normal tissue. The diagnosis of molecular genetics and cytogenetics prefers chromosome structure for analysis. Following the development of single cell techniques, a new term, “Single Cell Diagnosis”, has emerged in molecular pathology and molecular genetics/cytogenetics[30]. As shown in Table 5, single cell diagnosis can be utilized for several fields, such as pathology, genetics and microbiology. Now, this technique can be used in many clinical laboratories, for example, surgical specimens from operation, biopsy specimen from internal medicine (especially in haematology), blood samples from paediatrics (perinatal diagnosis),obstetrics/gynaecology and psychiatrics as well[31].
3.2.2. Forensic DNA analysis via Single Cell DNA-profiling
- The characteristics and the inheritance of autosomal and sex chromosomes are the basis to determine DNA typing in forensic medicine. Forensic DNA analysis, such as, restriction fragment length polymorphisms (RFLP), short tandem repeats (STR), variable number tandem repeats (VNTR), and mitochondrial DNA has been routinely applied into forensic laboratory. Along with human genome decoded and development of forensic-DNA techniques, the application of forensic-DNA analysis have extended from a blood sample into DNA trace specimen such as single cells or small number of cells. In RFLP technology, the polymorphism can be read by HPLC and DNA-Sequencer in a specific gene or non-coding DNA sequence in a individual person[32]; STR analysis combined with PCR technology with fluorescent labels, can automatically detect at a single cell level and analyzed the DNA genomic profile for a great amount of specimens[33]; because VNTR are inherited from the individual’s parents,, it also can be used for a single cell DNA-profiling[34]; Since mitochondrial DNA is passed from one generation to the next solely through the maternal line of a family, By comparing the mitochondrial DNA (mDNA)from samples of maternal families and maternal relatives, forensic scientists are able to search and demonstrate a specimen trace in a maternal family[35,36].
3.3. Clinical Treatment
3.3.1. Biomarker Discovery
- Early diagnosis and treatment is a key factor required to reduce the mortality and morbidity of all types of diseases, especially for tumor disease. Unfortunately, currently available cancer and genetic screening tools (CT, X-ray, mammography and invasive needle or surgical evaluation for cancer and genetic disease) are not sensitive enough for early detection of the diseases thus most tumor diseases cannot be treated at an early stage. Moreover, it is imperative to develop non-invasive technique for diseases, such as tumors that can be distinguished between patients with and without cancer, as well as stages of cancer. At present, genomic and proteomic technologies have rapidly been involved in cancer research[37, 38, 39]. Genomic technologies have allowed us to monitor thousands of gene expression profiles and evaluate functions of candidate genes to obtain a global view of cancer tissue. Proteomic techniques have also allowed us to understand proteins and their modifications. Despite its remarkable usefulness, microarray and proteomics techniques all have technical limitations because of cell purity or limited cell number after initial purification from clinical specimens so that, if we do not have a rational module for clinical genomic analysis, clinical genomic data cannot clearly define some specific biomarkers and the accompanied therapeutic targeting. In order to address the important question, here I introduce characteristics, advantages and disadvantages of the genomic analysis ex vivo, in vitro and in silico for application of clinical specimen. The most useful protocol uncovering biomarker is in vitro genomic analysis, such as single-cell manipulation by laser capture microscopy (LCM) along with genomic analysis. If we have known cell morphology in a tumor disease and/or information from cancer cell such as biomarker or telomerase activity, we can pick the cells with the morphological feature and enzyme activity, and then process genomic analysis from the harvested cells. After in vitro genomic analysis, we will uncover very specific biomarkers[40]. If the cells are located in liquid such as leukemia cells in bone marrow or peripheral blood, living cell-sorting such as FACS/MACS with downstream genomic analysis allows us to discover specific biomarkers from clinical specimens. Additionally, genomic analysis ex vivo, or living cell-sorting technique with the downstream cell culture to increase cell number or understand differentiating stage and then genomic analysis can help us to discover some biomarkers at different stages of cancer[41]. As mentioned above, genomic analysis ex vivo has been employed into cancer stem cell. Recently, genomic analysisin silico has been extensively utilized to discover tumor biomarker and biomarkers related with genetics. As shown in our result[42], genomic analysis in silico can screen or mine some very specific biomarkers in some diseases. Because of mixed different cells from clinical tissue remaining in the mined gene profile, current results from bioinformatic analysis still need further experiments, such as rtPCR or Western blot to confirm the mined gene profiles. Genomic analysis in silico is a good substitute method if the specimens have not been processed for micro-dissection in vitro orcell-sorting in/ex vivo.
3.3.2.Therapeutic Targeting
- Therapeutic targeting is a specific treatment for clinical diseases. In details, after genomic analysis along with pathway information and linking drug database to provide information, the corresponding drugs will directly target the explored therapeutic protein and nucleic acid for the targeted disease. A necessity of therapeutic targeting, clinical genomic analysis, nowadays, has been quickly applied into tumor disease, immune disease and genetic disease. Major developments are in two fields: A. therapeutic targeting for cancer stem cell; B. individualized therapy for metastatic tumor, immune disease or genetic disease.Now we all know, small populations of cells, called cancer stem cells (CSCs) located in tumor tissues play an important function in the development and progression of the disease[43,44,45]. It is also thought that CSCs drive the metastatic spread of cancer. CSCs are able to resist conventional therapies so that the disease is difficult to be completely eradicated. If therapeutic targeting identification can uncover some specific proteins or nucleic acid of CSCs, the selective targeting of CSCs will offer a new paradigm in both cancer diagnostics and therapeutics. According to current report, more than 30 CSC R&D program are subject to several clinical research to discover drugs or compound. Now, most CSC R&D program is being taken forward to large pharmaceutical companies[46,47]. In clinical fields, personalized medicine (or individualized therapy), one of special therapeutic targeting, is going to extend into different clinical diseases. Personalized medicine is a new medical model to be directly defined as physicians enable tailored approaches to prevent and care for individual patient relying on genomics and proteomics. It is often defined as "the right treatment for the right person at the right time." All examples of successful personalized treatments require a rational clinical genomic analysis. Based on research and development of clinical genomic analysis, we have successfully established a bioinformatics module for personalized therapy, that is, following genomic signature mined by genomic analysis in silico, quantitative pathway analysed by topology process, a specimen with no small cell lung cancer (NSCLC) was used to mine significant targeting, finally uncover therapeutic targeting with linking drug database for the special treatment[48].
4. Conclusions of Clinical Genomic Analysis
- Seven years ago, I describe single cell genomic techniques based on experiences in our laboratory and other laboratories. After further research and development of clinical genomic techniques with maturity in amplification for genomic DNA/RNA and signal magnification for protein for the past several years, many new genomic techniques have been developed into clinical specimens, for instances, dynamically single-cell or small number of cell-sorting techniques with downstream ex vivo culture to increase the cell number or differentiation into a special type of cells and then genomic analysis; moreover, genomic analysis at tissue level with in silico analysis has also be quickly developed. These new clinical genomic analyses have been extended from molecular pathology, molecular genetics/cytogenetics, forensic medicine into stem cells for regenerative medicine and tumor cells and/or cancer stem cells for biomarker discovery and therapeutic targeting and into T-cell for adoptive immunotherapy.
References
| [1] | R. Kuppers, H. Kanzler, M. L. Hansmann, K. Rajewsky. K. “Single cell analysis of Hodgkin/Reed-Sternberg cells,” Ann Oncol, vol. 7, pp.27-30, 1996. |
| [2] | R. Steinert, T. Buschmann, M. van der Linden, L. M. Fels, H. Lippert, M. A. Reymond. “The role of proteomics in the diagnosis and outcome prediction in colorectal cancer,”Technol Cancer Res Treat. vol 1, pp. 297-304, 2002. |
| [3] | C. A. Klein. “From single disseminated tumor cells to metastasis insights from molecular genetic analyses of single cells,” Verh Dtsch Ges Pathol. vol. 87, pp. 158-164, 2003. |
| [4] | A. Bernheim. “Cytogenetics, cytogenomics and cancer,”Bull Cancer. vol. 89, pp. 161-165, 2002. |
| [5] | R. Pallini, L. Ricci-Vitiani, G. L. Banna, M. Signore, D.Lombardi, M. Todaro, G. Stassi, M. Martini, G. Maira, L. M. Larocca, R. De Maria.“Cancer stem cell analysis and clinical outcome in patients with glioblastoma multiforme,” Clin Cancer Res. vol. 14, pp. 8205-8212, 2008. |
| [6] | H. Schmid, A. Henger, C. D. Cohen, K. Frach, H. J. Grone, D. Schlondorff, M. Kretzler. “Gene expression profiles of podocyte-associated molecules as diagnostic markers in acquired proteinuric diseases, J Am Soc Nephrol. vol. 14, pp. 2958-2966, 2003. |
| [7] | H. Lahdesmaki, L. Shmulevich, V. Dunmire, O. Yli-Harja, W. Zhang. “In silico microdissection of microarray data from heterogeneous cell populations,” BMC Bioinformatics. vol. 6, pp.54-57, 2005. |
| [8] | R. A. Edwards. “Laser capture microdissection of mammalian tissue,” J Vis Exp. vol. 8, pp.309-313, 2007. |
| [9] | K. Chiu, H. T. Lau, K. F. So, R. C. Chang. “Micro-dissection of rat brain for RNA or protein extraction from specific brain region,” J Vis Exp. vol. 7, pp269-275, 2007. |
| [10] | H. S. Erickson, J. W. Gillespie, M. R. mmert-Buck. “Tissue microdissection,” Methods Mol Biol. Vol. 424, pp. 433-448, 2008. |
| [11] | B. Li. “A strategy to identify genomic expression profiles at single-cell level and a small number of cells (review paper),” Journal of Biotechnology, vol. 7, pp71-82, 2008. |
| [12] | M. Ormerod, “Flow Cytometry: A practical approach,” Oxford University Press, Oxford, UK. 2000. |
| [13] | S. S. Wang, W. Kamphuis, I. Huitinga, J. N. Zhou, D. F. Swaab. “Gene expression analysis in the human hypothalamus in depression by laser microdissection and real-time PCR: the presence of multiple receptor imbalances,” Mol Psychiatry, vol. 1, pp. 86-799, 2008. |
| [14] | Z. N. Demou. “Time-lapse analysis and microdissection of living 3D melanoma cell cultures for genomics and proteomics,” Biotechnol Bioeng, vol. 101, pp.307-316, 2008. |
| [15] | B. Li, J. Ding, A. Larson, S. Song. “Tumor Tissue Recycling-A new combination therapy for solid tumor: experimental and preliminarily clinical research,” Anticancer (IN VIVO) vol. 13, pp.1-6, 1999. |
| [16] | J. E. Dick, G. Guenechea, O. Gan, C. Dorrell. “In vivo dynamics of human stem cell repopulation in NOD/SCID Mice,” Annals of the New York Academy of Sciences, pp.184-190, 1999. |
| [17] | U. Gurok, R. W. Loebbert, A. H. Meyer, R. Mueller, H. Schoemaker, G. Gross, B. Behl. “Laser capture microdissection and microarray analysis of dividing neural progenitor cells from the adult rat hippocampus,” Eur J Neurosci. vol. 26, pp. 1079-1090, 2007. |
| [18] | Y. Niyaz, M. Stich, B. Sägmüller, R. Burgemeister, G. Friedemann, U. Sauer, R. Gangnus, K. Schütze. “Noncontact laser microdissection and pressure catapulting: sample preparation for genomic, transcriptomic, and proteomic analysis,” Methods Mol Med. vol. 114, pp. 1-24, 2005. |
| [19] | M. R. Emmert-Buck, R. F. Bonner, P. D. Smith, R. F. Chuaqui, Z. Zhuang, S. R. Goldstein, R. A. Weiss, L. A. Liotta. “Laser capture microdissection,” Science, vol. 274, pp.998-1001, 1996. |
| [20] | M. Vandewoestyne, D. Van Hoofstat, F. Van Nieuwerburgh, D. Deforce. “Automatic detection of spermatozoa for laser capture microdissection,” Int J Legal Med. vol. 2, pp169-175, 2009. |
| [21] | T. C. Lund, L. B. Anderson, V. McCullar, L. Higgins, G. H. Yun, B. Grazywacz, M. R. Verneris, J. S. Miller. “iTRAQ is a useful method to screen for membrane-bound proteins differentially expressed in human natural killer cell types,” J Proteome Res. vol. 6, pp.644-653, 2007. |
| [22] | F. E. Koehn. “High impact technologies for natural products screening,” Prog Drug Res, vol. 65, pp.77-210, 2007. |
| [23] | W. Zhang, J. Ding, Y. Qu, H. Hu, M. Lin, A. Datta, A. Larson, G. E. Liu, B. Li. “Genomic expression analysis by single-cell mRNA differential display of quiescent CD8 T cells from tumour-infiltrating lymphocytes obtained from in vivo liver tumours,” Immunology, vol. 127, pp.83-90, 2009. |
| [24] | H. Lähdesmäki, L. Shmulevich, V. Dunmire, O. Yli-Harja, W. Zhang. “In silico microdissection of microarray data from heterogeneous cell populations,” BMC Bioinformatics. vol. 6, pp. 54-58, 2005. |
| [25] | P. G. Spetsieris, Y. Ma, V. Dhawan, D. Eidelberg. “Differential diagnosis of parkinsonian syndromes using PCA-based functional imaging features,” Neuroimage. vol. 45, pp.241-1252, 2009. |
| [26] | S. Y. Tuoya, H. Satoh, , D. Yu, Y. Matsuura, H. Tokutaka, M. Seno. “Spherical self-organizing map as a helpful tool to identify category-specific cell surface markers,” Biochem Biophys Res Commun. vol. 376, pp414-418, 2008. |
| [27] | R. P. Lanza, R. S. Langer, J. P. Vacanti. “Principles of Tissue Engineering, 2nd Ed.San Diego,” Academic Press. pp. 444, 2000. |
| [28] | J. A. Thomson, J. Itskovitz-Eldor, S. S. Shapiro. “Embryonic stem cell lines derived from human blastocysts,” Proc Natl Acad Sci USA, vol. 282, pp. 1145-1147, 1998. |
| [29] | L. S. Meirelles and N. B. Nardi. “Methodology, biology and clinical applications of mesenchymal stem cells,” Front Biosci. Vol. 14, pp. 4281-4298, 2009. |
| [30] | D. S. Zarlenga and J. Hinggins. “PCR as a diagnostic and quantitative technique in veterinary parasitology,” Vet Parasitol. vol. 101, pp70-80, 2000. |
| [31] | M. Harz, M. Kiehntopf, S. Stöckel, P. Rösch, E. Straube. T. Deufel, J Popp. “Direct analysis of clinical relevant single bacterial cells from cerebrospinal fluid during bacterial meningitis by means of micro-Raman spectroscopy,” J Biophotonics. vol. 2, pp. 70-80, 2009. |
| [32] | S. H. Yeung, T. S. Seo, C. A Crouse, S. A. Greenspoon, T. N. Chiesl, J. D. Ban, R. A. Mathies. “Fluorescence energy transfer-labeled primers for high-performance forensic DNA profiling,” Electrophoresis, vol. 29, pp. 2251-2259, 2008. |
| [33] | R. Chakraborty, D. N. Stivers, B. Su, Y. Zhong, B. Budowle. “The utility of short tandem repeat loci beyond human identification: implications for development of new DNA typing systems,” Electrophoresis, vol. 20, pp, 682-1696, 1999. |
| [34] | R. de Santis, A. Ciammaruconi, G. Faggioni, R. D'Amelio, C. Marianelli, F. Lista. “Lab on a chip genotyping for Brucella spp. based on 15-loci Multi Locus VNTR Analysis,” BMC Microbiol. vol. 9, pp66-72, 2009. |
| [35] | R. S. Arnold, C. Q. Sun, J. C. Richards, G. Grigoriev, I. M. Coleman, P. S. Nelson, C. L. Hsieh, J. K. Lee, , Z. Xu, A. Rogatko, A. O. Osunkoya, M. Zayzafoon, L. Chung, J. A. Petros. “Mitochondrial DNA mutation stimulates prostate cancer growth in bone stromal environment,” Prostate vol.69, pp. 1-11, 2009. |
| [36] | A. M. de Azeredo-Espin and A. C.Lessinger. “Genetic approaches for studying myiasis-causing flies: molecular markers and mitochondrial genomics,” Genetica vol. 126, pp. 111-131, 2006. |
| [37] | S. J. Cordwell, A. S. Nouwens, N. M. Verrills, D. J. Basseal, B. J. Walsh. “Subproteomics based upon protein cellular location and relative solubilities in conjunction with composite two-dimensional electrophoresis gels,” Electrophoresis, vol. 21, pp. 1094-1103, 2000. |
| [38] | J. M. Levsky, S. M. Shenoy, R. C. Pezo, R. H. Singer. “Single-cell gene expression profiling,” Science vol. 297, pp. 836-840, 2002. |
| [39] | G. Liu, X. Yuan, Z. Zeng, P. Tunici, H. Ng , I. R. Abdulkadir, L. Lu, D. Irvin, K. L. Black, J. S. Yu . “Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma,” Mol Cancer, vol.5, pp. 6-7, 2006. |
| [40] | C. Wei, W. Guo-min, L. Yu-jun. “Apoptosis resistance can be used in screening the markers of cancer stem cells,” Med Hypotheses, vol. 67, pp.1381-1383, 2006. |
| [41] | R. Besançon, S. Valsesia-Wittmann, A. Puisieux, C. C. de Fromentel, V. Maguer-Satta. “Cancer stem cells: the emerging challenge of drug targeting,” Curr Med Chem. vol. 16, pp. 394-416, 2009. |
| [42] | J. Q. Ding, H. L. Hu, B. Li. ”Genomic analysis and diagnosis for clinical biopsy specimens--using microarray database combined with micro-dissection in silico,”Genome informatics. Cold Spring Harbor Laboratory, vol. 42, pp. 20, 2008. |
| [43] | J. E. Trosko. “Cancer stem cells and cancer non stem cells: from adult stem cells or from reprogramming of differentiated somatic cells,” Vet Pathol. vol. 46, pp. 176-193, 2009. |
| [44] | R. Amin and L. Mishra. “Liver stem cells and tgf-Beta in hepatic carcinogenesis,” Gastrointest Cancer Res. vol. 2, ppS27-30, 2008. |
| [45] | S. A. Stuart, Y. Minami, J. Y. Wang. “The CML stem cell: Evolution of the progenitor,” Cell Cycle. vol. 17, pp.14-18, 2009. |
| [46] | M. Santisteban, J. M. Reiman, M. K. Asiedu, M. D. Behrens, A. Nassar, K. R. Kalli, P. Haluska, J. N. Ingle, L. C. Hartmann, M. H. Manjili, , D. C. Radisky, S. Ferrone, K. L. Knutson. “Immune-induced epithelial to mesenchymal transition in vivo generates breast cancer stem cells,” Cancer Res. vol. 69, pp. 2887-2095, 2009. |
| [47] | H. J. Lenz. “Colon cancer stem cells: a new target in the war against cancer,” Gastrointest Cancer Res. vol. 2, pp.203-204, 2008. |
| [48] | B. Li, N. Senzer, D. Rao, A. W. ong, P. Kumar, P. B. Maples, J. J. Nemunaitis.Bioinformatics “Approach to Individual Cancer Target Identification,” 11th Annual Meeting of the American Society of Gene Therapy. vol.8, pp. 451004, 2008. |
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-Text HTML
Full-Text HTML