-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Chemistry
p-ISSN: 2165-8749 e-ISSN: 2165-8781
2021; 11(3): 43-48
doi:10.5923/j.chemistry.20211103.01
Received: Jun. 30, 2021; Accepted: Jul. 17, 2021; Published: Jul. 31, 2021

Synthesis, Structure Analysis and Antibacterial Activity of Zn(II) and Co(III) Complexes
Amnah Saud Altamimi1, Salma A. Al-Zahrani2, Violeta Jevtovic2
1Master Student at Department of Chemistry, University Hail, City, Hail, Kingdom of Saudi Arabia
2Department of Chemistry, University Hail, City, Hail, Kingdom of Saudi Arabia
Correspondence to: Amnah Saud Altamimi, Master Student at Department of Chemistry, University Hail, City, Hail, Kingdom of Saudi Arabia.
| Email: |  |
Copyright © 2021 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

This paper presents the synthesis, structure and biological activity of two new complexes Zn (II) and Co (III) with the pyridoxal-thiosemicarbazone as ligand. Complexes are with molecular formula [Zn (PLTSC) (H2O)2]SO4. H2O and [Co (PLTSC-H)2] Cl.H2O. The synthesized complexes were structurally characterized and biological activity was performed on several microorganisms. The zinc complex has the structure of a distorted square pyramid, while the bis-ligand cobalt complex is a regular octahedron.
Keywords: Pyridoxal-thiosemicarbazone, Zn (II) and Co (III) complexes, Structure, Synthesis and biological activity
Cite this paper: Amnah Saud Altamimi, Salma A. Al-Zahrani, Violeta Jevtovic, Synthesis, Structure Analysis and Antibacterial Activity of Zn(II) and Co(III) Complexes, American Journal of Chemistry, Vol. 11 No. 3, 2021, pp. 43-48. doi: 10.5923/j.chemistry.20211103.01.
Article Outline
1. Introduction
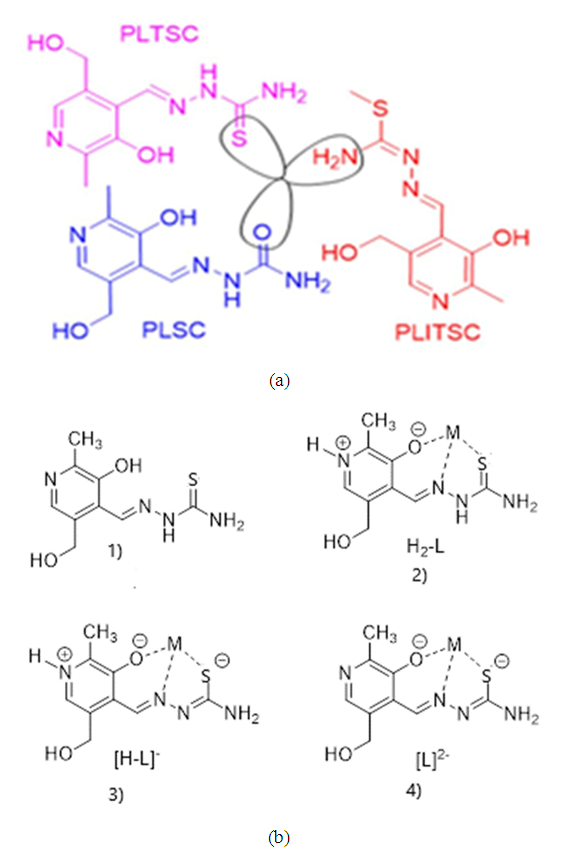
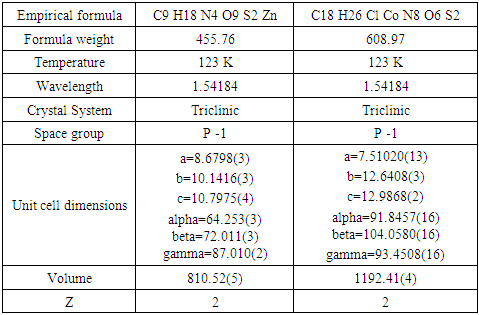
- Complexes with the ligand pyridoxal thiosemicarbazone (PLTSC) have been investigated so far and many papers [1,2] on the subject are known, including one book [3]. Pyridoxal- thiosemicarbazone (PLTSC) belongs to the family of ligand pyridoxal and carbazone derivatives, where, in addition to Pyridoxal- thiosemicarbazone (PLTSC) exist Pyridoxal-semi carbazone (PLSC) and Pyridoxal- S-methyl iso-thiosemicarbazone (PLITSC) (Scheme 1) are also known.All three ligands are tridentate. First place of coordination is oxygen from phenol hydroxyl, the second is Nitrogen from hydrazine group. Can be seen in Scheme 1 (a), for these three types of ligands differs is only third place of coordination with the central metal. Actually, for PLTSC it is a sulphur, for PLSC it is oxygen, while in ligand PLITSC in coordination is the NH group in the third place of coordination. Can be concluded that with manipulate its ligand backbone, lateral functionalities, and metal core to realize purpose specific ligands (Scheme 1 (b)). An added advantage of such ligands is that they can coordinate to the metallic core via different mode (Scheme 1 (b) (1-4)). For instance, it has been demonstrated that in complexes based on Pyridoxal semi-carbazones (PLSC) [3-11] and Pyridoxal S methyl iso-thiosemicarbazone (PLITSC) [12-18], metal can coordinate to the ligand via three different modes namely neutral (H2L), monoanionic (HL-) and dianionic (L2-) forms.
2. Experimental Part
- Commercial pyridoxal hydrochloride (PL·HCI, "Aldrich") and thiosemicarbazone (TSC, "Merck") were used for ligand synthesis.
2.1. Synthesis of Ligand PLTSC·H2O
- Solution 0.60g (3mmo1) PL·HCI was mixed with warm solution 0.27 g (3 mmol) TSC 8 cm3 H2O and it was poured into 2 cm3 H2O. The obtained yellow solution was stirred and left at room temperature for 10 hours. Deposited yellow acerate crystals were filtered off and washed two times with H2O. Yield: 0.60 g (69%).
2.2. Synthesis of [Zn (PLTSC) (H2O)2]SO4. H2O Complex
- 0.10 g (0.45 mmol) of the ligand PLTSC was dissolved in 15 cm3 of warm H2O and this solution was added 0.17 g (0.7 mmol) ZnSO4. The obtained yellow solution was left at room temperature around 3 days. Large yellow-orange crystals (monocrystals can also be found there) were filtered off and washed with H2O. Yield: 0.12 g (58%).
2.3. Synthesis of [Co (PLTSC-H)2] Cl.H2O Complex
- Mixture of 0.20 g (0.7 mmol) PLTSC·3H2O and 0.22 g (1 mmol) CoCl2 6H2O was perfused with 30 cm3 H2O. It was heated until complete dissolution of reactants and purple solution was left at room temperature for 24 hours. The obtained crystals were filtered off and dried in vacuum. Yield: 0.24 g (88%).
2.4. X-ray Crystallography
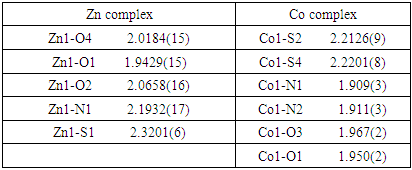
- A single crystal of 0.12 x 0.09 x 0.17 dimension was examined. The temperature was 296 K on a computing data collection For the X-ray measurements, single crystal of the complex was mounted on a glass fibber and examined at 296 K on a Bruker D8 Venture APEX diffractometer equipped with Photon 100 CCD area detector using graphite-monochromate Mo-Kα radiation [λ = 0.71073 Å]. In general, in the difference map the hydrogen atoms were 109 visible. Hydrogen atoms bound to carbon were initially positioned geometrically while the hydrogen atoms for the coordinated water molecules were located in the difference map. All hydrogen positions and isotropic displacement parameters were then refined in a separate cycle. Hydrogen positions were checked for feasibility by examination of the hydrogen-bonding network. Crystallographic data in the Cambridge Crystallographic Data Centre (CCDC, 12 Union Road, Cambridge CB2 IEZ, UK; e-mail: depos-it@ccdc.cam.ac.uk) were deposit, CCDC deposition number are 2092612 2092620. Crystal data collection and structure refinement are given in Table 1.
|
2.5. Bioassays
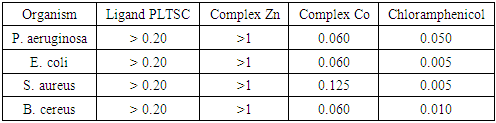
- In order to obtain the quantitative data for the reported compounds, the micro dilution technique was used (NCCLS, 2000) [23]. The following bacteria were tested: Escherichia coli (ATCC 10526), Pseudomonas aeruginosa (ATCC 27853), Staphylococcus aureus (ATCC 11632), and Bacillus cereus (ATCC 10876). Cultures of the test bacteria were growing overnight in Müller-Hinton agar at 37°C and then transferred to sterile saline. The bacterial suspension was adjusted with sterile saline to a concentration of approximately 1.0 × 107 cells/ml. The 1 ml of bacterial suspension was homogenized with 9 ml of melted (45°C) Müeller-Hinton agar. Minimum inhibitory concentrations (MICs) determination was performed by a serial dilution technique, using 96-well microtiter plates. The inoculate applied contained approximately 1.0 × 105 cells in a final volume of 100 µl/well. The compounds tested were dissolved in H2O (compounds [Zn (PLTSC) (H2O)2]SO4. H2O and [Co (PLTSC-H)2] Cl H2O.; conc. 5 × 10-3/10 ml H2O) and added to the broth medium with bacterial inoculate in a volume of 100 µl/well. The covered plates were incubated under aerobic conditions at 37°C for 24 h. The lowest concentrations without visible growth (at the binocular microscope) were defined as concentrations which completely inhibited bacterial growth (MICs). DMSO was used as a negative control, while chloramphenicol was used as a positive control. Dilutions of the inoculate were also cultured on solid MH to verify the absence of contamination and to check their validity. All tests were performed in triplicate.
3. Results and Discussion
3.1. Zink Complex [Zn (PLTSC) (H2O)2]SO4. H2O
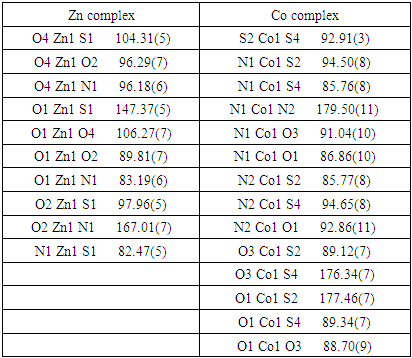
- The molecular structure for new complex of Zn is [Zn (PLTSC) (H2O)2]SO4. H2O shown in Figure 1. It crystallizes in P -1 space group with one discrete neutral unit [Zn (PLTSC) (H2O)2] and one SO4. anion and H2O molecule in out sphere. The environment around the central zinc atom can be best described as a distorted square-planar pyramid geometry. Scheme 2 shows the synthetic routes to the Zn (II) complex. The reactions of ZnSO4 with warm H2O solution of PLSC 2H2O, produced the title complex compound of Zn in the reaction of Zn salt with ligand the molar ratio was 1:1. 1992. Italian researchers managed to synthesize a complex formed in the reaction of ZnCl2 and PLTSC [19]. Reaction of zinc chloride or acetate with ligand, pyridoxal thiosemicarbazone (PLTSC), in that paper [19], lead to the formation of novel complexes which have been characterized by spectroscopic methods and X-ray analysis. That Zn complex is with formula [Zn (HL)Cl] 2H2O. The ligand PLTSC is in monoanionic form coordinated, the central metal Zn with two chlorine atoms in coordination also, so, the geometry is the square-pyramid.
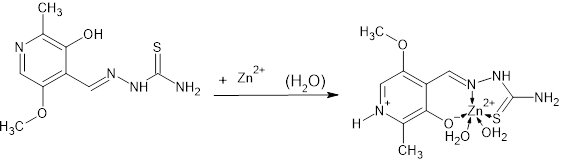 | Scheme 2. Synthesis of Zn complex compound |
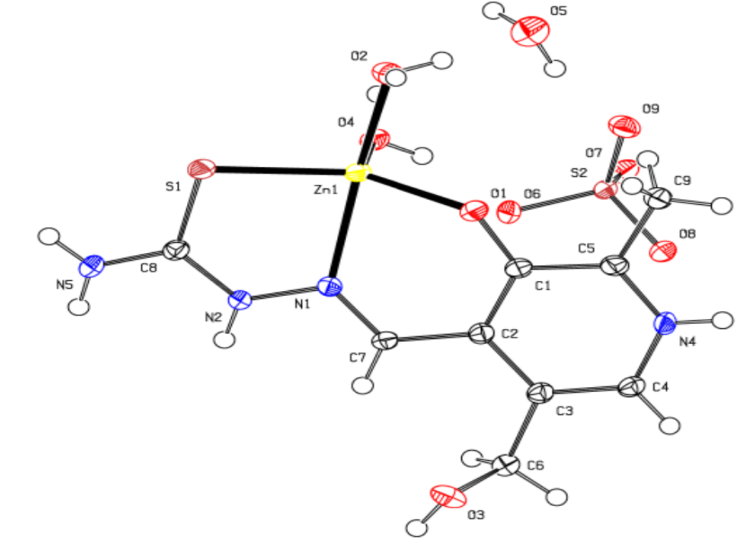 | Figure 1. The crystal structure for Zn complex |
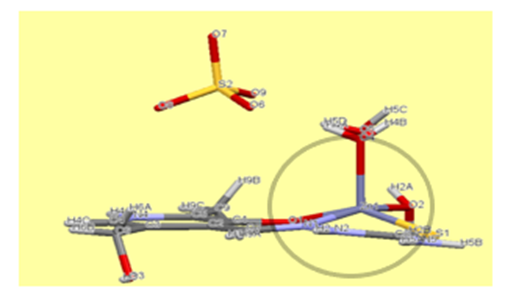 | Figure 2. A distorted square pyramid of Zn complex |
|
|
3.2. Cobalt Complex [Co (PLTSC-H)2] Cl.H2O
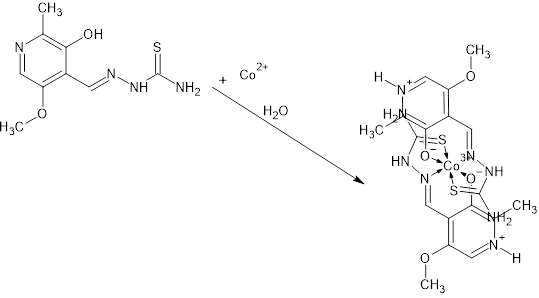
- As for the second complex, cobalt with PLTSC, the papers on that topic have been described so far [20,21]. Cobalt (III) Pyridoxal Thiosemicarbazone Complexes with Nitroprusside [20] is interesting from the aspect of pronounced biological activity, proving to be a good antileukemic agent.From the structural aspect, pyridoxal N (4)-substituted thiosemicarbazone cobalt (III) complex [20] is much more important for us. The structure of Cobalt in the work of Indian authors [20] is absolutely identical to our structure of cobalt, the only difference is in the existence of C6H6 molecule on the terminal nitrogen from the thiocarbazone chain. Actually, the investigation of effect of substitution (CH or C H) on terminal N (4)-nitrogen of thiosemicarbazone exhibited its influence on the potential binding and cleavage ability with DNA, BSA binding, free radical scavenging and cytotoxicity.Our cobalt (III) complex, perhaps thanks to a similar structure as the complex from the research of Indian authors [20], is also very biologically active, more precisely its antibacterial effect on several known bacteria has been investigated in this paper.The our [Co (PLTSC-H)2] Cl.H2O complex was obtained from the simple salt CoCl2, while the starting salt for the complex pyridoxal N (4)-substituted thiosemicarbazone cobalt (III) [20] was [CoCl2 (PPh3)2].The title cobalt complex is a bis-ligand complex, with the octahedral structure (Figure 3). The synthesis process is given in Scheme 3, an aqueous solution of cobalt (II) chloride salt is used which, in reaction with the PLTSC ligand in a ratio of 1: 2, gives purple crystals, stable in air.
 | Scheme 3. Synthesis of Co complex compound |
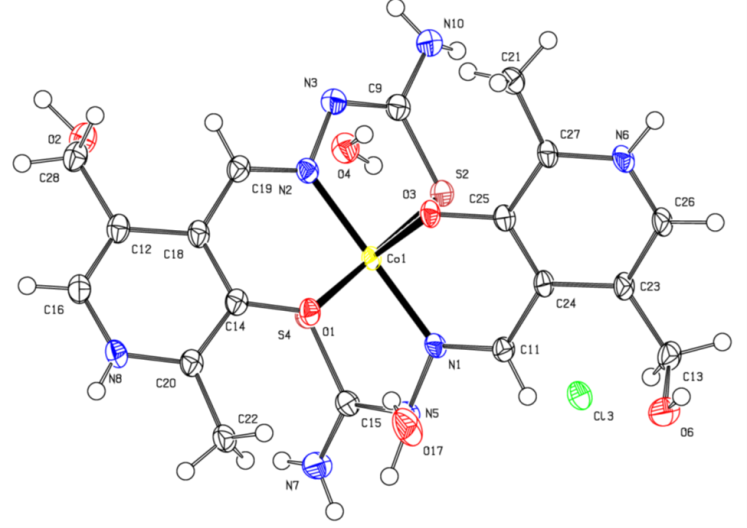 | Figure 3. Crystal Structure of Co complex |
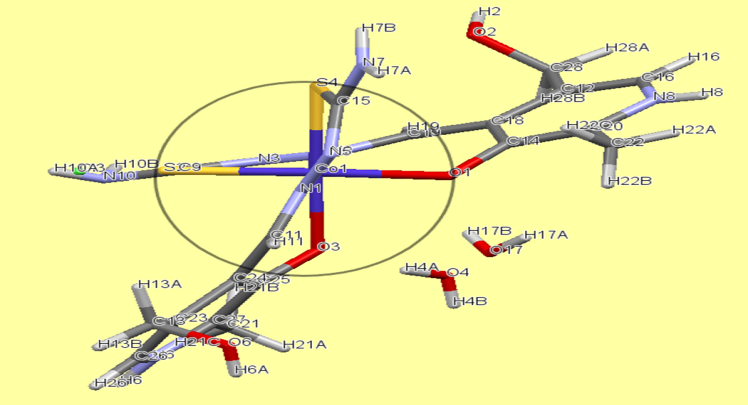 | Figure 4. Octahedral structure of Co complex |
3.3. Biological Activity
- In the Table 4 is given the summary of the antimicrobial activities of the ligand (PLTSC) and complexes (Zn and Co). As can see, the complex Co shows the greatest antibacterial activity in comparison to the other tested substances. The fact that complex Co showed activity towards both Gram negative (Escherichia coli, Pseudomonas aeruginosa) and Gram positive (Staphylococcus aureus, Bacillus cereus) bacteria. According that fact, can be concluded that the antibacterial mechanism of complex Co is not dependent on the structural features of the bacterial cell wall, what give high performance to Co complex like antibacterial compound.
|
4. Conclusions
- Two new complexes of Zinc and Cobalt were synthesized with the ligand Pyridoxal thiosemicarbazone (PLTSC), with formulas [Zn (PLTSC) (H2O)2]SO4. H2O and [Co (PLTSC-H)2] Cl.H2O. X-ray structural analysis showed that in the case of the zinc complex it is a monoligand complex, with coordination number 5, (ONS ligand PLTSC and 2 molecules of water in coordination with Zn (II) central metal), square pyramidal geometry. The cobalt complex is a bis-ligand complex (two ONS ligands PLTSC in coordination with Co (III) central metal, octahedral geometry. The syntheses for both complexes were performed in aqueous solution, with the corresponding Zn and Co salts (ZnSO4 and CoCl2), during which the oxidation of Co2+ to Co3+ occurred during the coordination reaction, while the oxidation number of zinc remained unchanged (+2) as in the initial salt ZnSO4.The complexes were biologically tested where they proved to be promising antibacterial agents. The cobalt complex has been shown to be more active than the zinc complex or the ligand itself. Surprisingly, its activity is enforceable regardless of whether it is gram-positive or negative bacteria, so it can be expected that its good therapeutic performance may find practical application in some of the further research.Actually, Gram-negative bacteria are surrounded by a thin peptidoglycan cell wall, which itself is surrounded by an outer membrane containing lipopolysaccharide. Gram-positive bacteria lack an outer membrane but are surrounded by layers of peptidoglycan many times thicker than is found in the Gram-negatives [25].Based on this, it is little unexpected that our results show the best activity in the case of Gram-positive Staphylococcus aureus. In the case of Gram-positive Bacillus cereus, such an effect would also be absent, since these are positive bacteria in both cases. In any case, without a doubt, the tests should be expanded and cover as many different types of bacteria as possible, but also other types of tests should be done, anticancer, for example.
ACKNOWLEDGEMENTS
- Author Amna Saud Altamimi, a Master student at University Hail, along with her mentors, would like to thank University Hail for its support and opportunities to use the university's laboratory services. Also, she expresses great gratitude for the analyzes done at the University of Monash, Victoria, Australia, especially to Dr. Dragoslav Vidovic and his research group of PhD students, who helped a lot in creating this research work.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML