-
Paper Information
- Next Paper
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Chemistry
p-ISSN: 2165-8749 e-ISSN: 2165-8781
2014; 4(1): 42-46
doi:10.5923/j.chemistry.20140401.07
Synthesis, Characterization and X-ray Analysis of a Copper(II) Complex with Pyridoxal-aminoguanidine Ligand
Violeta Jevtovic
Department of Chemistry, College of Sciences, University of Sharjah, Sharjah, United Arab Emirates
Correspondence to: Violeta Jevtovic, Department of Chemistry, College of Sciences, University of Sharjah, Sharjah, United Arab Emirates.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
The reaction between pyridoxal hydrochloride (PL•HCl) and aminoguanidine-hydrogencarbonate (AG.H2CO3) in the presence of Na2CO3•10H2O resulted in the formation of pyridoxal-aminoguanidine (PLAG; H2L) ligand. The ligand’s coordination chemistry with CuCl2 yielded a green-brown, octahedral dimmer of Cu (II) with the general formula of [Cu(PLAG)(NCS)2]2. This paper presents the synthesis and the structural analysis of this complex with the Schiff-based ligand.
Keywords: Pyridoxal-aminoguanidine Cu(II) complex, Synthesis, Crystal structure, Characterization
Cite this paper: Violeta Jevtovic, Synthesis, Characterization and X-ray Analysis of a Copper(II) Complex with Pyridoxal-aminoguanidine Ligand, American Journal of Chemistry, Vol. 4 No. 1, 2014, pp. 42-46. doi: 10.5923/j.chemistry.20140401.07.
Article Outline
1. Introduction
- Hyperglycemia has been identified as a major risk factor for the development of diabetic complications[1]. Aminoguanidine (AG) has been extensively studied as one of the most promising compounds for the treatment of diabetic complications, because the compound has both advanced glycation inhibitory activity[2] and antioxidant activity[3,4]. It has been reported that a pyridoxal- aminoguanidine (PL-AG) Schiff base adduct exhibits advanced glycation inhibitory activity comparable to that of AG, while causing no decrease in the liver pyridoxal phosphate content of normal mice[5,6]. Also, PL-AG is more potent than AG in preventing nephropathy in streptozotocin-indluced diabetic mice[7]. These findings suggest that PL-AG is superior to AG for the treatment of diabetic complications because not only it prevents vitamin B6 deficiency, but it is also superior at controlling diabetic nephropathy. The preventive effect of this adduct against diabetic nephropathy is mediated via inhibition of both oxidation and glycation[8]. The inhibition of advanced glycation end products (AGEs) at millimolar concentrations of AGEs inhibitors, used in many in vitro studies, results primarily from the copper chelating or antioxidant activity of the AGEs inhibitors, rather than their carbonyl trapping activity[9]. Defining the chelating activity of AGEs inhibitors is essential for understanding the mechanism of action of drugs and possible benefits of chelation therapy in diabetes, as well as for developing more effective clinically useful inhibitors of diabetic complications. Bearing in mind the biological and medicinal importance of AG and PL-AG adduct and the influence of copper (II) ions on their inhibitory activity, in the present study evaluation of the chelating ability of PL-AG inhibitor was attempted in order to cast more light on the role of copper chelation in the mechanism of action of AGEs inhibitors.
2. Experimental Section
2.1. Reactants
- All commercially obtained reagent-grade chemicals were used without further purification, except for the ligand, which was prepared according to the previously described procedure[10a,b].
2.2. Physical Measurements
- Elemental (C, H, N) analysis of air-dried samples was carried out by standard micromethods inthe Center for Instrumental Analysis, Faculty of Chemistry, Belgrade. The X-ray analysis was performed in the Oxford Chemical Crystallography Service.
2.3. Crystal Data Collection and Processing
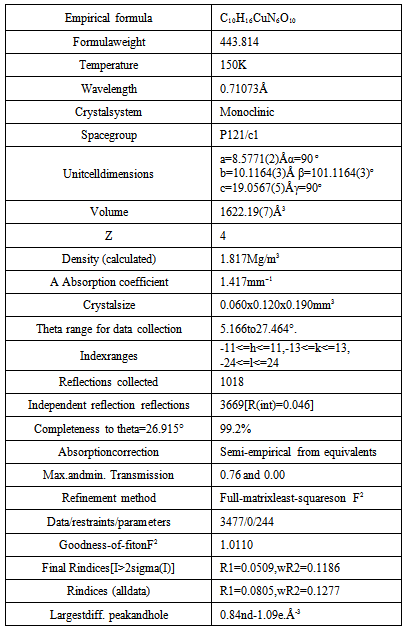
- A single crystal was mounted on a glass fibberin random orientation. Data collection was performed at temperature 150 K on a computing data collection Nonius Kappa CCD diffractometer using graphite mono-chromated MoKα radiation (λ = the data can be obtained free of charge by writing to 0.71073 Ǻ). Data collection and cell refinement we details concerning data collection and structure re carried out using DENZO and COLLECT[11,12]. The structures were solved with SIR-92 and refined using CRYSTALS [13,14]. In general, the hydrogen atoms were visible in the difference map. Therefore, they were positioned geometrically and refined in a separate hydrogen cycle (with soft restraints) before inclusion in the refinement using a riding model. For more information see the Supplementary information. Crystallographic data has been deposited at the Cambridge Crystallographic Data Centre. Copies of thedata can be obtained free of charge by writing to CCDC, 12 Union Road, Cambridge CB2 IEZ, UK; e-mail:deposit@ccdc.cam.ac.uk. The CCDC deposition numbers are 828036. Crystal data anddetails concerning data collection and structure refinement are given in Table 1.
|
2.4. Preparation of the Ligand and Complex [Cu(PLAG)(NCS)2]2
2.4.1. PLAG•HCl•H2O
- The ligand (PL-AG•HCl•H2O), synthesized in a similar manner as described for PL-AG[6], is a yellow microcrystalline compound which is moderately soluble in water and sparingly soluble in MeOH, EtOH and DMF[10].
2.4.2. [Cu(PLAG)(NCS)2]2
- To a warm methanol (20cm3) solution containing 0.28g (1 mmol) of PLAG 0.26g (1.2 mmol) of CuCl2 in 10cm3MeOH was added followed by 0.12 g (1.6 mmol) of NH4NCS. Green mixture was formed which was left behind at room temperature for about 50 hours. Green crystals started to form after a couple of hours. After the sedimentation has completed the product was isolated by vacuum filtration and was then washed with methanol. Yield: 0.18 g.
3. Results and Discussion
3.1. Synthesis and Characterization of the Ligand and the Complex
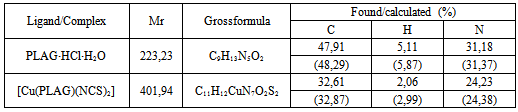
- Neutral form of the ligand pyridoxal-aminoguanidine (PLAG), is synthesized according to a previously described procedure[10], involving a reaction of water mixture between PL•HCl and AG•H2CO3[1:1] in the presence of Na2CO3•10H2O (Scheme 1).PLAG is a yellow microcrystalline compoundvery soluble in MeOH and DMF, but slightly less in H2O and EtOH. The resulting compound is stable in air and at high temperatures. The elemental analysis data are given in Table 2.By using reaction of a warm MeOH mixture of CuCl2 and PLAG, the mole ratio of 1:1, with the addition of NH4NCS a dark-green monocrystaldimeric complex[Cu(PLAG) (NCS)2]2 (Scheme 2) was obtained.
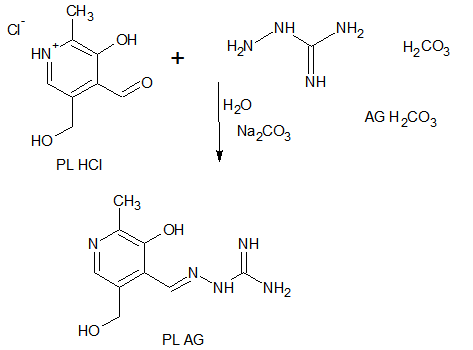 | Scheme 1. Synthesis of pyridoxal-aminoguanidine (PLAG) |
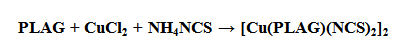 | Scheme 2. Syntheses of the complex[Cu (PLAG)(NCS)2]2 |
|
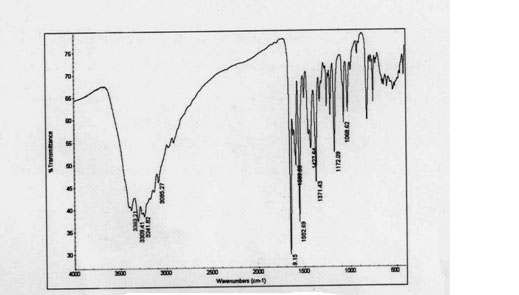 | Figure 1. IR spectra of complex |
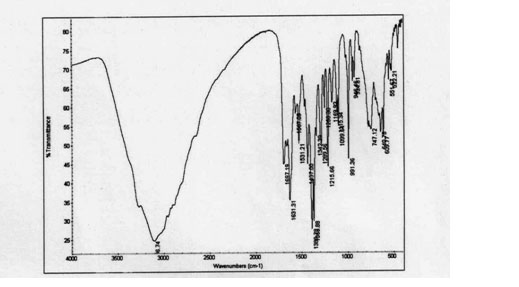 | Figure 2. IR spectra of PLAG |
3.2. X-ray Crystallography
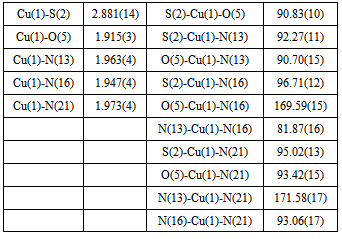
- As shown in the empirical formula, the complex contains a neutral form of PLAG ligand. The neutral form represents a proton transfer from the phenolic hydroxyl to the pyridine nitrogen atom. The doubly deprotonated form occurs from deprotonation of the phenolic oxygen and hydrazine nitrogen atoms, similar to a series of ligands withpyridoxalcarbazones[15].The complex[Cu(PLAG)(NCS)2]2 possesses a hexacoordi-nated copper ion formed with the usual tridentate ONN coordination of ligand PLAG, coordination of two NCS-groups and a bonding interaction with the nearby Cu atom from a different molecule (Figure 3). Chelating ligand coordination is achieved through the oxygen atom of deprotonated phenolic hydroxyl, azomethine group nitrogen atom and guanido group nitrogen atom. As it can be seen from Table 1, the complex crystallizes in the monoclinic system, with four dimer molecules (Z=4) in the unit cell. Coordination sphere of the complex was built from the neutral form of the ligand molecule PLAG and two NCS groups. In this complex, as already mentioned, the tridentate ONN ligand in its neutral form is coordinated by building two metallocycles: one six-membered (pyridoksilydene) and one five-membered (aminoguanidine).The bond lengths of for the metal-ligand distances and the corresponding bond angles are given in Table 3.
|
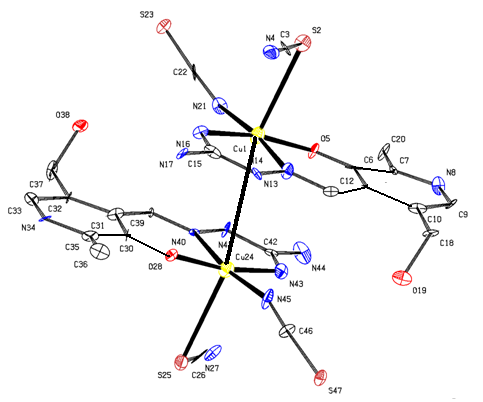 | Figure 3. Crystal structure of [Cu(PLAG)(NCS)2]2 |
4. Conclusions
- The synthesis, characterization and structural analysis of a Cu(II) complex incorporating PLAG ligand is described. The Cu II environment is best described as octahedral. The equatorial plane is formed by the tridentate ONN-coordinated PLAG ligand and one NCS molecule. This compound crystallizes in a monoclinic crystal symmetry with the space group of P 1 21/c 1.
5. Supplementary Information
- Crystallographic data (excluding structure factors) for the structure (1) reported in this paper have been deposited with the Cambridge Crystallographic Data Center, CCDC No. 828036. Copy of the data can be obtained free of charge on application to CCDC, 12 Union Road, Cambridge CB21EZ, UK(fax:+441223336033;e-mail:deposit@ccdc.cam.ac.uk http://www.ccdc.cam.ac.uk).
ACKNOWLEDGMENTS
- The author acknowledge the Oxford Chemical Crystal-log-raphy Service for the use of the instrumentation and many thanks DJWatkin research group for help.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML