-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Chemistry
p-ISSN: 2165-8749 e-ISSN: 2165-8781
2013; 3(5): 148-152
doi:10.5923/j.chemistry.20130305.05
Synthesis, Characterization and X-ray Crystal Structure of the Dimer Complex[Cu (PLITSC-2H) (NH3)]2·2H2O
Violeta Jevtovic
Department of Chemistry, College Sciences, University of Sharjah, Sharjah, United Arab Emirates
Correspondence to: Violeta Jevtovic, Department of Chemistry, College Sciences, University of Sharjah, Sharjah, United Arab Emirates.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
The title complex[Cu(ITSCPL-2H)(NH3)]2.2H2O (1), is reported. The structure of the title compound, C20 H34 Cu2 O6 N10 S2 as an interesting metal complex with a Schiff base derived from isothiosemicarbazide and pyridoxal (3-hydroxy-5-hydroxymethyl-2-methylpyridine-4-carboxaldehyde) is reported. Ligand pyridoxal-S-methyliso thiosemicarbazone (PLITSC; H2L) is tridentate ONN ligand. The CuII environment is a square planar coordination, the equatorial plane of which is formed by the tridentate ONN-coordinated pyridoxal-Smethyliso thiosemicarbazone and one amonium molecule. This compound crystallizes in triclinic symmetry, in space group P-1, with lattice constans: a = 9.0161(2)Ǻ, b = 10.9078(3)Ǻ, c = 16.6355(5)Ǻ, α = 105.1276(12)°, β = 90.9785(10)°, γ = 111.0567 (13)°, V = 1462.68(7) Å3, Z = 3, F(000) = 742, R1 = 0.0452, wR2 = 0.1110. The crystal structure, molecules are linked by O-H…O, N-H…O and C-H…O contacts. The complex was characterized by elemental analysis, IR spectra, conductometric and magnetochemical measurements, and X-ray diffraction.
Keywords: Pyridoxal-S-methyliso thiosemicarbazone, Ni (II) complex, Synthesis, Characterisation, Crystal structure
Cite this paper: Violeta Jevtovic, Synthesis, Characterization and X-ray Crystal Structure of the Dimer Complex[Cu (PLITSC-2H) (NH3)]2·2H2O, American Journal of Chemistry, Vol. 3 No. 5, 2013, pp. 148-152. doi: 10.5923/j.chemistry.20130305.05.
Article Outline
1. Introduction
- Transition metal complexes with ligand based on thiosemicarbazides (TSC) have been studied for many years because of their interesting structural properties and biological activities[1,2]. The complexes with TSC-based ligands exhibit a wide range of biologically important properties, such as antiviral, antitumor and anti-inflammatoryactivities[3,4]. Isothiosemicarbazones (ITSC) can also act as biologically active antibacterial agents[5]. There are many complexes incorporating the pyridoxal-S-methyliso thiosemicarbazone (PLITSC) ligand in its tridentate ONN coordination mode and the most important results gained in studying these types of complexes have been reviewed by Leovac[6]. Almost all of the reported complexes including several Fe-base compounds such as [Fe(ITSCPL)(ITSCPL-H)](NO3)3, [Fe(ITSCPL)Cl3]·H2O and[Fe(ITSCPL)2]OAc·2H2O[7,8] were characterized by physical properties (elemental (C,H,N) analysis, molar conductivities) and their voltametric properties. It is worth noting that the syntheses and characterizations of several of these complexes were also included as a topic of a PhD thesis[9]. However, the only X-ray characterized complex that incorporates PLITSCligand is [CuII(NO3)(PLITSC)(H2O)] (NO3)a and [Ni(PLITSC)(H2O)3](NO3)2b[10]. The PLITSC ligand acts as a tridentate ligand in this CuII complex forming two fused chelate rings, one being five-membered (ITSC) and one six-membered (PL). The PLITSC ligand in the title complex 1 is coordinated in the same manner as the reported for the CuII complex.
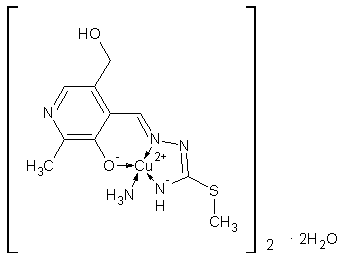 | Scheme 1. Complex 1 |
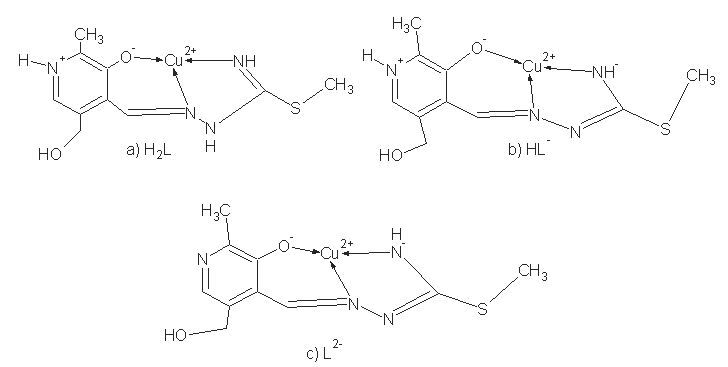 | Scheme 2. Coordination models and ligand forms: a) neutral, b) mono- and c) dianionic |
2. Experimental Section
2.1. Physical Measurements
- All commercially obtained reagent-grade chemicals were used without further purification, except for the ligands, which were prepared according to the previously described procedures[8]. Elemental (C,H,N) analysis of air-dried samples was carried out by standard micromethods in the Center for Instrumental Analysis, Faculty of Chemistry, Belgrade. Magnetic susceptibilities were measured at room temperature using a magnetic susceptibility balance MSB-MKL (Sherwood Scientific Ltd. Cambridge, England). Molar conductivities of the freshly prepared 110-3 M solution were measured on a Jenway 4010 conductivity meter. IR spectra (KBr disk) were recorded on a Thermo Nicolet (NEXUS 670 FT-IR) instrument. The X-ray analysis was performed in Oxford Chemical Crystallography Service.
2.2. Preparation of the Title Compound
- The pyridoxal 3-methylisothiosemicarbazone ligand, H2L, was prepared by the reaction of an ethanolic solution of 3-methylisithiosemicar-bazidehydroiodide with pyridoxal hydrochloride, and subsequent neutralisation with an aqueous solution of Na2CO3. Brown single crystals of 1 were obtained by the reaction of a hot methanol solution of the ligand with CuSO4.5H2O (molar ratio 1:1). NH3 should be intercalated into stirred solution in the drops until it becomes transparent. Yield: 74%. μeff = 1.32 BM; elemental analysis calcd. (%) for C20H34Cu2N10O6S2: C 33.37, H 5.04, N 19.46; S 8.91; found: C 33.38, H 5.09, N19.96; S 8.53. λ M(DMF)= 1.6 Scm 2 mol-1. IR (KBr, Nujol) Suporting material.
2.3. X-ray Crystallography
- A single crystal of 0.01 x 0.01 x 0.01 was mounted on a glass fiber in a random orientation. Data collection was performed at temperature 150 K on a computing data collection Nonius Kappa CCD diffractometer using graphite monochromated MoKα radiation (λ = 0.71073Ǻ). Data collection and cell refinement were carried out 106 using SIR92[12] and DENZO and COLLECT[13]. The structure of 107 compound 1 was solved with SIR-92 and refined using 108 CRYSTALS[14, 15]. In general, the hydrogen atoms were 109 visible in the difference map. Hydrogen atoms bound to carbon were initially positioned geometrically while the hydrogen atoms for the coordinated water molecules were located in the difference map. All hydrogen positions and isotropic displacement parameters were then refined in a separate cycle, using restraints prior to inclusion into the model with riding constraints. Hydrogen positions were checked for feasibility by examination of the hydrogen-bonding network. Crystallographic data have been deposited at the Cambridge Crystallographic Data Centre. Copies of the data can be obtained free of charge by writing to the CCDC, 12 Union Road, Cambridge CB2 IEZ, UK;e-mail:depos-it@ccdc.cam.ac.uk. The CCDC deposition number is 754806.Crystal data and details concerning data collection and structure refinement are given in Table 1.
|
3. Results and Discussion
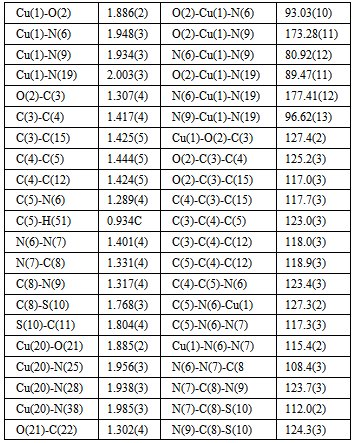
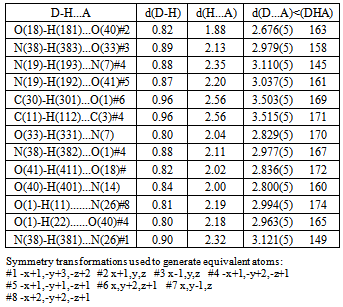
- Regarding the majority of Cu(II) complexes (which are coloured green), complex is distinguishable by its characteristically dark colour. Cu(II) complexes have similar characteristics (colour) with tridentate ONN salicylaldehyde S-methylisothiosemicarbazone (H2L) as well, which are green, with the exception of Cu(L)NH3.H2O which is dark-coloured[16]. Complex 1 has reduced μeff (1.32) value, which can be explained by its dimeric structure, most often realized via oxygen atoms of phenolic hydroxyl. These oxygen atoms function as bridges[17] in case of tridentate ONN isothiosemicarbazones.The examined complex is badly dissoluble. It can be dissolved in DMF, while λM(DMF) = 1.6 value indicates its non-electrolytic nature[18].IR spectrum of[Cu(ITSCPL-2H)(NH3)]2.2H2O complex has no prominent bands[19] at about 2800cm-1, which can be understood, as regards deprotonation of pyridine nitrogen atom.X-ray analysis of[Cu(ITSCPL-2H)(NH3)]2.2H2O (1), showed a square planar structure, formed by the tridentate ONN coordination of the PLITSC ligand and one NH3 molecules.The title compound crystallizes in the space group P-1. The molecular structure and atom-numbering scheme are show in Fig.1.The analysis of the crystal structure of 1 has been carried out and experimental details are presented, whereas its geometrical features (bond distances and angles) are given in Table 2 and in Table 3 are given possible hydrogen bonds.
|
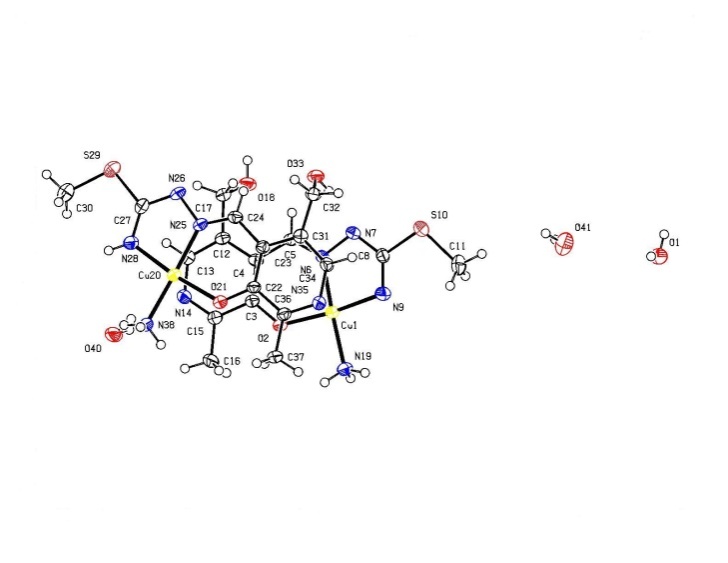 | Figure 1. Complex 1 |
|
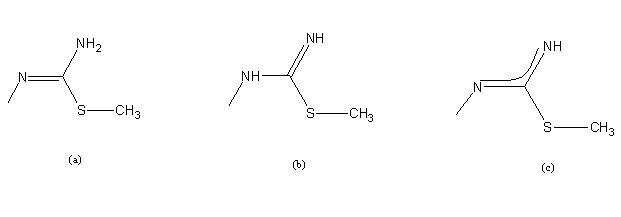 | Scheme 3. Amido (a), imido- (b) and deprotonated (c) forma of ITSC |
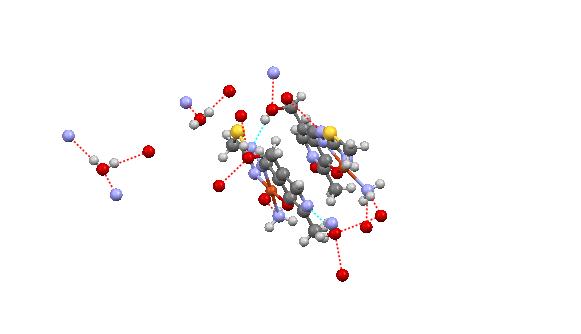 | Figure 2. Packing arrangement between two asymmetric units and several intermolecular hydrogen bonding interactions |
4. Conclusions
- The synthesis, characterisation and structural analysis of Cu complexes incorporating PLITSC ligand is described. The CuII environment is a square planar coordination, the equatorial plane of which is formed by the tridentate ONN-coordinated PLITSC ligand and one ammonium molecule. This compound crystallizes in triclinic symmetry, in space group P-1.
5. Supplementary Information
- Crystallographic data (excluding structure factors) for the structure (1) reported in this paper have been deposited with the Cambridge Crystallographic Data Center, CCDC No. 754806. Copy of the data can be obtained free of charge on application to CCDC, 12 Union Road, Cambridge CB2 1EZ, UK (fax: +44 1223 336033; e-mail: deposit@ccdc.cam.ac.uk or http://www.ccdc.cam.ac.uk).
ACKNOWLEDGMENTS
- The author acknowledge the Oxford Chemical Crystallography Service for the use of the instrumentation and many thanks DJWatkin research group for help.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML