-
Paper Information
- Next Paper
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Chemistry
p-ISSN: 2165-8749 e-ISSN: 2165-8781
2012; 2(3): 157-159
doi: 10.5923/j.chemistry.20120203.10
Palladium Complex Containing Curcumin as Ligand: Thermal and Spectral Characterization
Mônica Aparecida Rodrigues, Jehorgyelly Nunes Fernandes, Reinaldo Ruggiero, Wendell Guerra
Chemistry Institute, Federal University of Uberlândia, João Naves de Ávila Avenue, 2121, Campus Santa Mônica, Uberlândia - MG, 38.400-902, Brazil
Correspondence to: Wendell Guerra, Chemistry Institute, Federal University of Uberlândia, João Naves de Ávila Avenue, 2121, Campus Santa Mônica, Uberlândia - MG, 38.400-902, Brazil.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
Reaction of curcumin with K2PdCl4 at pH 9.0 (60℃) produces a neutral complex of palladium containing curcumin as ligand. The compound was characterized by conductivity measurements, elemental analysis, infrared and UV-visible electronic spectroscopy and thermogravimetric analysis (TGA/DTA). The spectroscopic techniques show that the ligand is coordinated to palladium(II) via -diketone group. Thermal studies supported the chemical formulation of this complex and showed that the same has general structure [Pd(Cur-H)2].2H2O.
Keywords: Palladium Complexes, Curcumin, Metal Dye Complexes
1. Introduction
- Curcumin, a component of Curcuma Longa rhizomes is a phenolic compound that has a series of beneficial properties such as antitumor activity, anti-inflammatory and antioxidant[1-5]. This series of beneficial properties has caused an explosive growth in the interest of the academic community to understand how this compound acts, for example, in preventing cancer. Thus, a series of studies are ongoing, including phase of clinical trials, against a great number of diseases such as cancer, psoriasis and Alzheimer's disease[5-9].In the biological conditions, curcumin can coordinate with metallic ions through the acetylacetonate group. In almost all work reported in literature, complexes of curcumin formed is of the type ML2 where the metallic ion has electrical charge +2, and complex of the type ML3, when the ion metallic has electrical charge +3[10-15]. Many these complexes were evaluated for various biological activities and very interesting results have been reported[15-21]. An interesting example refers to a palladium complex containing bypyridine and curcumin, that induces both cell growth inhibition as apoptosis of human prostate cancer cells (LnCaP, PC3, and DU145), through the production of ROS and JNK phosphorylation associated with GSTp1 down regulation[22]. Curiously, palladium complex containing only the curcumin has not been studied and, it would be interesting, since this neutral compound can be more active and useful.These observations encouraged us to furtherinvestigations. Thus, in the present study; we report the thermal and spectral characterization of complex of palladium containing curcumin. Subsequently, their biological properties will be evaluated.
2. Results & Discussion
- A palladium complex containing curcumin as ligand was synthesized using a methodology different of the described in literature for [Pd(Curc-H)2]4Ace[23]. The structure this compound was proposed considering only elemental analysis data. In this work, the complex containing curcumin, [Pd(Curc-H)2].2H2O, was characterized by elemental analysis, conductivity measurements, IR, UV-Vis and thermogravimetric analysis. The chemical structure of the ligand and their palladium complex are presented in Figure 1.The results of the elemental analyses (C, H, N and Pd) are in accordance with the proposed structure.The molar conductance measurement of the palladium complex was performed in DMF solution (with a concentration of 1×10-) at room temperature. The value obtained (4.02 S·cm2 ·mol-1) indicates that the compound is nonelectrolytes[24].
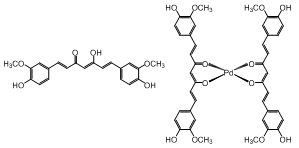 | Figure 1. Structure of curcumin and of palladium complex obtained |
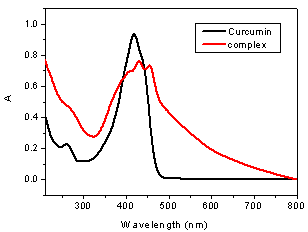 | Figure 2. Electronic spectrum of the curcumin and its palladium complex (acetonitrile, 1.0 x 10-5 mol L-1) |
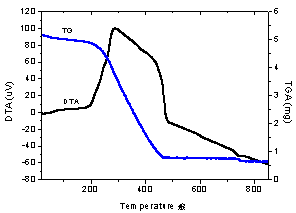 | Figure 3. TGA/DTA curves for palladium complex |
3. Conclusions
- A complex of palladium was isolated with curcumin and characterized by spectroscopic and thermals techniques. The results show that the palladium ion bind to curcumin occurs via the oxygen atoms of the -diketone group. Thermal studies supported the chemical formulation of this complex and showed that the same decompose in various steps. Considering the structure of compound obtained, the investigation of its biological properties is desirable to determine their possible utility as drug, thus, later their properties of cytotoxity and antioxidant will be performed in our laboratory.
4. Experimental
- Starting materialsThe reagents are commercially available (Aldrich). All other reagents chemicals were of analytical grade, purchased from different sources, and used without further purification.Physical measurementsConductivity studies were carried out with a Digimed DM 31 conductivity meter using a cell of constant 1.00 cm-1, spectroscopic grade dimethylformamide (Merck) (M = 1.20 μs/cm-1) and tetraethylammonium bromide (M = 78.39 μs/cm-1) as a standard.Elemental analyses (C, H and N) were performed using a Perkin-Elmer 2400 CHN Elemental Analyser. Palladium content was determined by atomic absorption on a Hitachi spectrophotometer model 8200.IR spectra were registered in KBr pellets on a Shimadzu FTIR-Irprestige-21 spectrometer. Spectrophotometer UV-2501 PC Shimadzu was used for UV and visible absorption measurements.Thermogravimetric analyses (TG/DTA) were obtained on a TGA-50 Shimadzu, using 6.0 mg samples packed in aluminum crucible. Samples were heated at 10℃/min from room temperature to 900℃, in a dynamic nitrogen atmosphere (flow rate = 200 mL/min). The residue was analyzed by X-ray diffratometry on a Siemens D5000 apparatus using a copper tube and radiation Cu Ka = 1.54178 Å, with 2 angle varying from 0 to 90°.Synthesis of complex of K2PdCl4 (0.5 mmol) previously dissolved in water was added to 5 mL of a methanolic solution of curcumin (1.0 mmol) at 60℃. After 2h, the pH was then adjusted to 9.0 by adding triethylamine dropwise and the mixture was stirred for 24h. The solid formed was separated by filtration, washed with methanol and water, and dried under vacuum.Complex -[Pd(Curc-H)2].2H2OYield: 65 %. Color: Brown. Anal. Calcd. for [Pd(C21H19O6)2]2H2O: C, 57.45; H, 4.78 %; N, 0%; Pd, 12.13 Found: C, 57.31; H, 4.56 %; N, 0%; Pd, 12.06. IR spectra in KBr, ν (cm-1): 3500, 2931, 2840, 1623, 1597, 1511, 1460, 1427, 1391, 1275, 1214, 1166, 1125, 1030, 971, 845, 820, 850, 724, 613, 564.
ACKNOWLEDGEMENTS
- This work was supported by grants of CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil) and FAPEMIG (Fundação de Amparo à Pesquisa de Minas Gerais, Brazil).
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-Text HTML
Full-Text HTML