-
Paper Information
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
International Journal of Biophysics
2012; 2(1): 7-11
doi: 10.5923/j.biophysics.20120201.02
A Theoretical Study on the Interaction of Thionine with Micrococcus Lysodeikticus DNA
Ghazala Yunus , Seema Srivastava , Vishwambhar D. Gupta
Department of Physics, Integral University, Kursi Road, Lucknow, 226026, India
Correspondence to: Ghazala Yunus , Department of Physics, Integral University, Kursi Road, Lucknow, 226026, India.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
The latest study carried out on binding of small molecules with double stranded deoxyribonucleic acid recommended that the intercalation of thionine, a tricyclic heteroaromatic molecule, with natural double stranded deoxyribonucleic acid granted thermal stabilization to the thionine-DNA complex. In this investigation, we reported theoretical analysis of thionine binding with Micrococcus lysodeikticus DNA duplex (Type XI, 72 mol% GC) by using a modified Zimm and Bragg theory, to elucidate the melting behaviour and heat capacity of Micrococcus lysodeikticus DNA with and without thionine binding. For implementing present study, the experimental models of Paul et al.(2010) have been used. The sharpness of transition has been examined in terms of half width and sensitivity parameter (∆H/σ). The conclusion of theoretical analysis recommended that the different parameters, for example, transition profile, heat capacity curve, sharpness of the transition and half widths are in good agreement with the experimental measurements for interaction of thionine. Thus, the theoretical analysis proposed in this study may be useful to understand interaction of small molecules to deoxyribonucleic acid that may be applied for the designing of prospective DNA binding therapeutic molecules and in the process of drug development.
Keywords: Thionine, Micrococcus Lysodeikticus DNA, Transition Profile, Heat Capacity, Drug Development
Article Outline
1. Introduction
- In the recent years the study of small molecules interaction with deoxyribonucleic acid is an investigation area of high tropical interest. There are number of techniques dedicated to support drug-DNA interactions which are increasing continuously. Over the past several years, the interaction of aromatic heterocyclic molecules to nucleic acids has received significant interest due to their relevance in a variety of biological applications including cancer chemotherapy[1-7]. Thus, a number of experimental studies are conducted to understand the nature and thermodynamics of heterocyclic aromatic molecules and nucleic acid interaction[6],[8-14]. A tricyclic heteroaromatic molecule, Thionine (3,7-Diamino-5- phenothiazinium) (Fig. 1), has been studied for its intercalative interaction with DNA duplex[10] and photoinduced mutagenic actions on binding to DNA[12]. According to Chang et al.[15] thionine binds specifically with guanine–cytosine (GC) contents of DNA duplex as suggested by satellite hole spectroscopy study. Although, Tuite and Kelly[10] suggested that (GC) specificity of thionine binding was not very marked. The other model recommended that thionine exists in two different tautomeric forms and the amino form is intercalated while the imino form is not[16-17]. These results provide somewhat contradicting data on the GC specificity of thionine binding to linear double stranded DNA. The findings of another study carried out through pressure tuning hole burning spectroscopy has conclusively revealed an external stacking mode of thionine binding with quadruplex structures[16]. Accordingly, although some structural data is available, the same is not conclusive and the thermodynamics of thionine binding to nucleic acid is not yet elucidated. Recently the calorimetric analysis of thionine binding to a DNA duplex along with the structural and thermodynamic aspects of thionine binding to natural DNAs of varying base composition has been reported by Paul et al.[6]. They concluded strong binding of thionine with Micrococcus lysodeikticus DNA (ML-DNA) which increases the thermal stability of the duplex, and at saturation the duplex melts with 9.4°C above that of the free duplex[6]. They also concluded that thionine binding to ML-DNA is an exothermic process and there is involvement of multiplicity of non covalent interactions in the binding process. The present study deals with theoretical analysis of thionine binding to understand the effect of its binding with a native DNA duplex using the experimental model of Paul et al.[6] who studied the thermal and thermodynamic behaviour of thionine binding to ML-DNA. The modified Zimm and Bragg theory, initially considered for helix coil transitions in polypeptides, are used to explain lambda point anomalies in heat capacities and order-disorder transition in thionine bounded and unbounded ML-DNA duplexes,[18-20]. The alteration in nucleation parameter, which is an inverse measure of binding strength, reflects the effect of thionine binding.
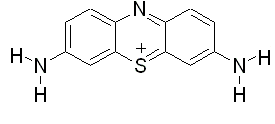 | Figure 1. Chemical structure of thionine |
2. Theory
- On the basis of spectroscopic and calorimetric study for the complex formation between thionine and DNA, as mentioned above, Paul et al.[6] recommended that phenothiazinium dye, thionine, binds robustly with the Micrococcus lysodeikticus DNA that resulted in thermal stabilization of the complex. The binding of thionine to the GC rich ML-DNA was intercalative and was non-cooperative that resulted in significant perturbation of the conformation of the DNA as well as thermal stabilization[6]. Although, the system remains a highly co-operative one hence the co-operative transition theory could be applied to elucidate the melting profile and temperature dependence of thermodynamical parameter, such as heat capacity. Thus, we used modified Zimm and Bragg theory[18] which is explained in our earlier publication[21]. In brief, the above mentioned theory consists of writing an Ising matrix for a two-phase system, the bounded state and unbounded state. As discussed previously[19-20],[22-24] and by Zimm and Bragg[18], the Ising matrix M can be written as:
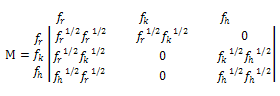 | (1) |
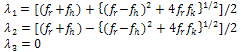 | (2) |
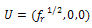 | (3) |
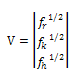 | (4) |
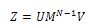 | (5) |
 | (6) |
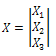 By substituting the values of M from Eq. (6), we get:
By substituting the values of M from Eq. (6), we get: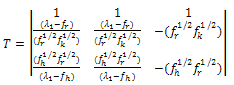 | (7) |
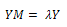 | (8) |
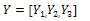 Again by substituting the values of M from Eq. (1) in Eq. (8), we get:
Again by substituting the values of M from Eq. (1) in Eq. (8), we get: 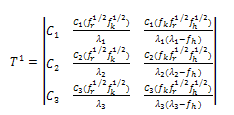 | (9) |
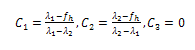 | (10) |
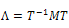 be the diagonalized form of M, the partition function can be written as:
be the diagonalized form of M, the partition function can be written as: 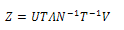 | (11) |
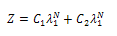 | (12) |
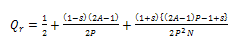 | (13) |
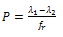 Here s is propagation parameter, which for simplicity is assumed to be 1. In fact, in most of the systems, it is found to be close to unity. If Ar and Ah are the absorbance in disordered and ordered states, respectively, the total absorption can be written as:
Here s is propagation parameter, which for simplicity is assumed to be 1. In fact, in most of the systems, it is found to be close to unity. If Ar and Ah are the absorbance in disordered and ordered states, respectively, the total absorption can be written as: 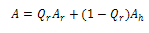 | (14) |
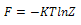 and
and 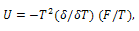 respectively. Differentiating internal energy with respect to temperature we get the specific heat:
respectively. Differentiating internal energy with respect to temperature we get the specific heat: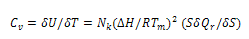 | (15) |
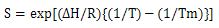 , Tm is the transition temperature, and
, Tm is the transition temperature, and
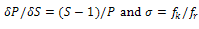 ; σ is the nucleation parameter and is a measure of the energy expanded/released in the formation (uncoiling) of first turn of the ordered/disordered state. It is related to the uninterrupted sequence lengths[18]. The volume heat capacity Cv has been converted into constant pressure heat capacity Cp by using the Nernst-Lindemann approximation[22]:
; σ is the nucleation parameter and is a measure of the energy expanded/released in the formation (uncoiling) of first turn of the ordered/disordered state. It is related to the uninterrupted sequence lengths[18]. The volume heat capacity Cv has been converted into constant pressure heat capacity Cp by using the Nernst-Lindemann approximation[22]: 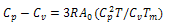 | (16) |
3. Results and Discussion
3.1. Transition Profiles
- The findings of spectroscopic and fluorescence studies, for thionine binding affinity with ML-DNA duplex, suggested that there is strong intercalative binding between thionine and ML-DNA and enabled the hypothesis of two state systems consisting of free and bounded thionine[6]. When a phenothiazinium dye, thionine, binds with GC rich natural ML-DNA duplex, the structure of DNA duplex still remains a very much co-operative and accordingly two-state theory of order-disorder transition is applicable. The Zimm and Bragg[18] theory is amended so as to consider ordered (bounded/unbounded) and disordered states as the two states which co-exist at the transition point. The transition is described mainly by the nucleation parameter and overall change of enthalpy/entropy, which are also the main thermodynamic forces driving the transition. The change in enthalpy obtained from differential scanning calorimetric (DSC) measurements takes all this into account. This is evident from the enthalpy change and changes in other transition parameters, such as nucleation parameter (σ) and melting point (Table 1). The results obtained from theoretical study suggested that the binding of thionine increases melting temperature of ML-DNA duplex at saturation point. At saturation, the melting point of DNA duplex was increased with 9.7°C in comparison to free duplex. The sharpness of the transition can be looked at in terms of half width and a sensitivity parameter defined as (∆H/σ). The deviations in half width and sensitivity parameter scientifically revealed that the transition is sharp in case of unbounded state and goes blunt with thionine saturation. In case of λ-transition, the same trend in the sharpness of transition is seen between the thionine bounded as well as unbounded curves. As anticipated, the sharpness is better in unbounded state, as compared to bounded state. A range of parameters, which give transition profiles in best agreement with the experimental measurements for binding of thionine to ML-DNA duplex, are given in Table 1.
3.2. Heat Capacity
- Heat capacity, second derivative of the free energy, has been calculated by using equation (14) which is used to characterize conformational and dynamical states of a macromolecular system. These heat capacity curves with λ-point anomaly and along with their transition profile are shown in figure 2. The results revealed that the theoretically obtained heat capacity profile agreed with the experimentally reported ones and could be brought almost into concurrence with the use of scaling factors, which is very close to one in transition profiles and slightly higher for the heat capacity curves. The sharpness of the transition can be characterized by the half width of the heat capacity curves that are in good agreement in both experimental and theoretical graphs.
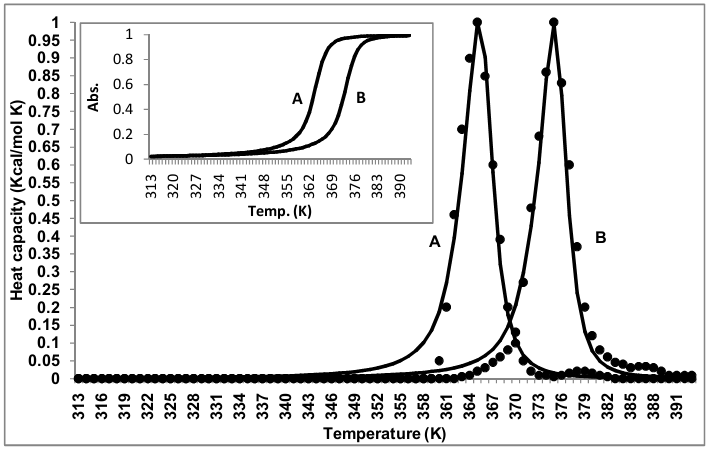 | Figure 2. Heat capacity and transition profiles (inset) for thionine bounded and unbounded ML-DNA. (A) Unbounded state, (B) Bounded state at saturation. [(—) calculated and (••••) experimental values] |
4. Conclusions
- The present study concluded that the natural Micrococcus lysodeikticus DNA molecule is an extremely co-operative structure and when thionine bind to it the co-operativity is not so much disturbed. Consequently, the modified Zimm and Bragg theory can be successfully applied to it. It generates the experimental transition profile and λ-point heat capacity anomaly effectively. These results will allow us to evaluate the thermodynamic profile of the binding process. Our theoretical studies of heterocyclic aromatic molecules binding are being extended to other synthetic and natural DNA also. Therefore, the theoretical analysis presented in this study can be implicated to understand bimolecular interactions and may also be applied in biomedical industries for drug design and development.
ACKNOWLEDGEMENTS
- Technical assistance from Dr. Mohammed Kuddus, Department of Biotechnology, Integral University, Lucknow is gratefully acknowledged.
References
| [1] | L.H. Hurley, “Secondary DNA structures as molecular targets for cancer therapeutics”, Biochemical Society Transactions, Vol. 29, N0. 6, pp 692–696, 2001. |
| [2] | L.H. Hurley, “DNA and its associated processes as targets for cancer therapy”, Nature Reviews Cancer, Vol. 2, pp 188 – 200, 2002. |
| [3] | R. Martinez and L. Chacon-Garcia, “The search of DNA-intercalators as antitumoral drugs: what it worked and what did not work”, Current Medicinal Chemistry, Vol. 12, pp 127–151, 2005. |
| [4] | R. Palchaudhuri and P.J. Hergenrother, “DNA as a target for anticancer compounds: methods to determine the mode of binding and the mechanism of action”, Current Opinion in Biotechnololy, Vol. 18, No. 6, pp 497 – 503, 2007. |
| [5] | M. Maiti and G.S. Kumar, “Molecular aspects on the interaction of protoberberine, benzophenanthridine, and aristolochia group of alkaloids with nucleic acid structures and biological perspectives”, Medicinal Research Reviews, vol. 27, No. 5, pp 649–695, 2007. |
| [6] | P. Paul, M. Hossain, R.C.Yadav and G.S. Kumar, “Biophysical studies on the base specificity and energetics of the DNA interaction of photoactive dye thionine: Spectroscopic and calorimetric approach”, Biophysical Chemistry, Vol. 148, No. 1-3, pp 93– 103, 2010. |
| [7] | K. Bhadra and G.S. Kumar, “Therapeutic potential of nucleic acid-binding isoquinoline alkaloids: binding aspects and implications for drug design,” Medicinal Research Reviews, Vol. 31, No. 6, pp 821–862, 2011. |
| [8] | C.V. Kumar and E.H. Asuncion, “DNA binding studies and site selective fluorescence sensitization of an anthryl probe”, Journal of American Chemical Society, Vol. 115, No. 19, pp 8547-8553, 1993. |
| [9] | E.M. Tuite and J.M. Kelly, “New trends in photobiology: photochemical interactions of methylene blue and analogues with DNA and other biological substrates”, Journal of Photo chemistry and Photobiology, B: Biology, Vol. 21, pp 103–124, 1993. |
| [10] | E.M. Tuite and J.M. Kelly, “The interaction of methylene blue, Azure B, and thionine with DNA: formation of complexes with polynucleotides and mononucleotides as model systems”, Biopolymers, Vol. 35, pp 419–433, 1995. |
| [11] | S. Jockusch, D. Lee, N.J. Turro and E.F. Leonard, “Photo-induced inactivation of viruses: adsorption of methylene blue, thionine, and thiopyronine on beta bacteriophage”, Proceeding of National Academy of Sciences USA, Vol. 93, pp 7446–7451, 1996. |
| [12] | C. Dohno, E.D.A. Stemp and J.K. Barton, “Fast back electron transfer prevents guanine damage by photoexcited thionine bound to DNA” Journal of American Chemical Society, Vol. 125, pp 9586–9587, 2003. |
| [13] | J.Y. Du, X.H. Huang, F. Xu, Y.Y. Feng, W. Xing and T.H. Lu, “Spectral study on the interaction mechanism between thionine and calf thymus DNA”, Guang Pu Xue Yu Guang Pu Fen Xi, Vol. 25, No. 9, 1435-1448, 2005. |
| [14] | Y. Xu, L. Yang, X. Ye, P. He and Y. Fang, “Impedance-based DNA biosensor employing molecular beacon DNA as probe and thionine as charge neutralizer”, Electroanalysis, Vol. 18, pp 873 – 881, 2006. |
| [15] | T.C. Chang, Y.P. Yang, K.H. Huang, C.C. Chang and C. Hecht, “Investigation of thionine–DNA interaction by satellite hole spectroscopy”, Optics and Spectroscopy, Vol. 98, pp 716– 721, 2005. |
| [16] | C. Hecht, J. Friedrich and T.C. Chang, “Interactions of thionin with DNA strands: intercalation versus external stacking’, Journal of Physical Chemistry B, Vol. 108, pp 10241–10244, 2004. |
| [17] | C. Hecht, P. Hermann, J. Friedrich and T.C. Chang, “Thionin in a cyclodextrin nanocavity: measuring local compressibilities by pressure tuning hole burning spectroscopy”, Chemical Physics Letters, Vol. 413, No. 4-6, pp 335 – 341, 2005. |
| [18] | B.H. Zimm and J.K. Bragg, ”Theory of the phase transition between helix and random coil in polypeptide chains”, Journal of Chemical Physics, Vol. 31, pp 526-535, 1959. |
| [19] | S. Srivastava, V.D. Gupta, P. Tandon, S. Singh and S.B. Katti, “Drug binding and order-order and order-disorder transitions in DNA triple helices”, Journal of Macromolecular Science Physics, Vol. 38, pp 349-366, 1999. |
| [20] | S. Srivastava, S. Srivastava, S. Singh and V.D. Gupta, “Stability and transition in a DNA tetraplex: a model for telomeres”, Journal of Macromolecular Science Physics, Vol. 40, No. 1, pp 1-14, 2001. |
| [21] | G. Yunus, S. Srivastava and V.D. Gupta, “Stability of DNA binding with dipyrandium: A theoretical study”, International Journal of Physical Sciences, Vol. 6, No. 36, pp 8151 – 8156, 2011. |
| [22] | K.A. Roles, and B. Wunderlich, “Heat capacities of solid poly(amino acids). I. Polyglycine, poly(L-alanine), and poly(L-valine)”, Biopolymers, Vol. 31, pp 477-487, 1991. |
| [23] | N. Poklar, D.S. Pilch, S.J. Lippard, E.A. Redding, S.U. Dunham and K.J. Breslauer, “Influence of cisplatin intrastrand crosslinking on the conformation, thermal stability, and energetics of a 20-mer DNA duplex”, Proceeding of National Academy of Sciences USA, Vol. 93, pp 7606-7611, 1996. |
| [24] | S. Srivastava, I.A. Khan, S. Srivastava and V.D. Gupta, “A theoretical study of the stability of DNA binding with cis/trans platin”, Indian Journal of Biochemistry and Biophysics, Vol. 41, pp 305-310, 2004. |
| [25] | J. Yuan, W. Guo and E. Wang, “Oligonucleotide stabilized silver nanoclusters as fluorescence probe for drug-DNA interaction investigation”, Analitica Chimica Acta, Vol. 706, No. 2, pp 338-342, 2011. |
| [26] | D.L. Ma, D.S. Chan, P. Lee, M.H. Kwan and C.H. Leung, “Molecular modeling of drug-DNA interactions: virtual screening to structure-based design”, Biochimie, Vol. 93, No. 8, pp 1252-1266, 2011. |
| [27] | F. Araya, G. Huchet, I. McGroarty, G.G. Skellern and R.D. Waigh, “Capillary electrophoresis for studying drug-DNA interactions”, Methods, Vol. 42, pp 141-149, 2007. |
| [28] | M.M. Islam, S.R. Chowdhury and G.S. Kumar, “Spectroscopic and calorimetric studies on the binding of alkaloids berberine, palmatine and coralyne to double stranded RNA polynucleotides, Journal of Physical Chemistry B, Vol. 113, pp 1210-1224, 2009. |
| [29] | V. Gonzalez-Ruiz, A.I. Olives, M.A. Martin, P. Ribelles, M.T. Ramos and J.C. Menendez, “An overview of analytical techniques employed to evidence drug-DNA interactions. Applications to the design of genosensors”, In: Biomedical engineering, trends, research and technologies, M. A. Komorowska and S. Olsztynska-Janus (eds.), InTech, pp 65-90, 2011. |
| [30] | A. Kunwar, S. Emmanuel, U. Singh, R.K. Chittela, D. Sharma, S.K. Sandur and I.K. Priyadarsini, “Interaction of a curcumin analogue dimethoxycurcumin with DNA”, Chemical Biology and Drug Design, Vol. 77, pp 281–287, 2011. |
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-Text HTML
Full-Text HTML