-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Bioinformatics Research
p-ISSN: 2167-6992 e-ISSN: 2167-6976
2023; 12(1): 1-10
doi:10.5923/j.bioinformatics.20231201.01
Received: Sep. 26, 2023; Accepted: Oct. 20, 2023; Published: Oct. 23, 2023

In silico Identification of MicroRNAs Potentially Targeting the Low-density Lipoprotein Receptor-Related Protein 5 (LRP5) Transcript, a Crucial Factor for Osteoporosis Development and Other Genes of a Predictive Network of LRP5
Mrinmoyee Sengupta, Mitali Das
Laboratory of Molecular Cell Biology and Genetics, Department of Zoology, University of Gour Banga, Malda, West Bengal, India
Correspondence to: Mitali Das, Laboratory of Molecular Cell Biology and Genetics, Department of Zoology, University of Gour Banga, Malda, West Bengal, India.
| Email: |  |
Copyright © 2023 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Low-density lipoprotein receptor-related protein 5 (LRP5) is the key regulator of bone metabolism. Deregulation of this gene may cause different bone metabolic diseases such as osteopororsis characterized by low BMD. Deregulation of this gene includes mutations, single nucleotide polymorphisms, altered expression and several others. Like all other genes, LRP5 also works through a complex genetic network. This study focuses on a genetic network of LRP5 proposed by GeneMania, a bioinformatics tool. A list of microRNAs targeting LRP5 has been retrieved from miRDB database and validated through other software. In the proposed genetic network, 20 genes are found to be interacting with LRP5 by various means. The 22 miRNAs found to interact with LRP5 has also found to influence the other genes from the proposed genetic network of LRP5. A meta-analysis shows some of these miRNAs to play important role in bone metabolism as well as osteoporosis development. As miRNAs regulate gene expression through a complex regulatory network, the rest of the miRNAs also need experimental validations of their functional role in bone. Over all this paper is a comprehensive report of miRNAs affecting LRP5 function in bone development and diseases particularly osteoporosis.
Keywords: Bioinformatics, GeneMania, in silico, LRP5, miRDB, miRNA, Osteoporosis, Venny 2.1
Cite this paper: Mrinmoyee Sengupta, Mitali Das, In silico Identification of MicroRNAs Potentially Targeting the Low-density Lipoprotein Receptor-Related Protein 5 (LRP5) Transcript, a Crucial Factor for Osteoporosis Development and Other Genes of a Predictive Network of LRP5, American Journal of Bioinformatics Research, Vol. 12 No. 1, 2023, pp. 1-10. doi: 10.5923/j.bioinformatics.20231201.01.
Article Outline
1. Introduction
- The low-density lipoprotein receptor-related protein 5 (LRP5) is encoded by the LRP5 gene in humans [1]. LRP5 is the key component of the LRP5/LRP6/Frizzled co-receptor complex working through canonical Wnt signaling pathway regulating bone metabolism. LRP5 thus has an important role in bone metabolism as it transduces signals by Wnt proteins [2] in osteoblasts [3]. It is evident that any alteration in the expression of LRP5 can lead to considerable changes in bone mass following bone diseases [4].Osteopororsis is a complex bone metabolic disease characterized by low bone mineral density. It has been raised as a major public health hazard now a days among the elderly population [5]. Through recent investigations LRP5 gene is appearing as one of the key regulators of bone mineral density as well as of the development of osteoporosis [6].Lrp5 gene, located on human chromosome 11q13 [7] associated with several other genes to form a genetic network. Altered expression of LRP5 may affect the functionality of these genes and vice versa. MicroRNAs (miRNAs) are proved to be one of the most important regulators of eukaryotic gene expression [8]. The miRNAs are short single stranded non-coding RNAs usually 19-25 nucleotides long. They target and bind to messenger RNAs (mRNAs) and determine their fate [9]. Binding of miRNAs clog translation of target mRNAs into protein and encourage degradation of mRNA targets. In this way miRNAs regulate the expression of more than 30% of protein-coding genes at the post-transcriptional and translational level [10]. miRNAs have immense role in regulating LRP5 expression as well as its function. Several miRNAs reported to target LRP5 which are enlisted in various target prediction software. Comprehensive and computational analysis of miRNA targeting LRP5 as a crucial factor in osteoporosis progression and bone metabolism has not yet been undertaken. Aim of this piece of work is to assemble all the available information about miRNAs taking part in regulation of LRP5 and thus affecting its role in bone metabolism.
2. Materials and Methods
2.1. Prediction of Genetic Network of LRP5
- GeneMania (http://www.GeneMania.org) is an online bioinformatics tool which predicts the interaction of a number of genes with the query gene [11] using a wide set of functional association data. We put human “LRP5” of as our query term and retrieved a genetic interaction network of 20 genes associated with LRP5 [12].
2.2. Mining the miRNA Pool Targeting LRP5 and Its Interacting Partners
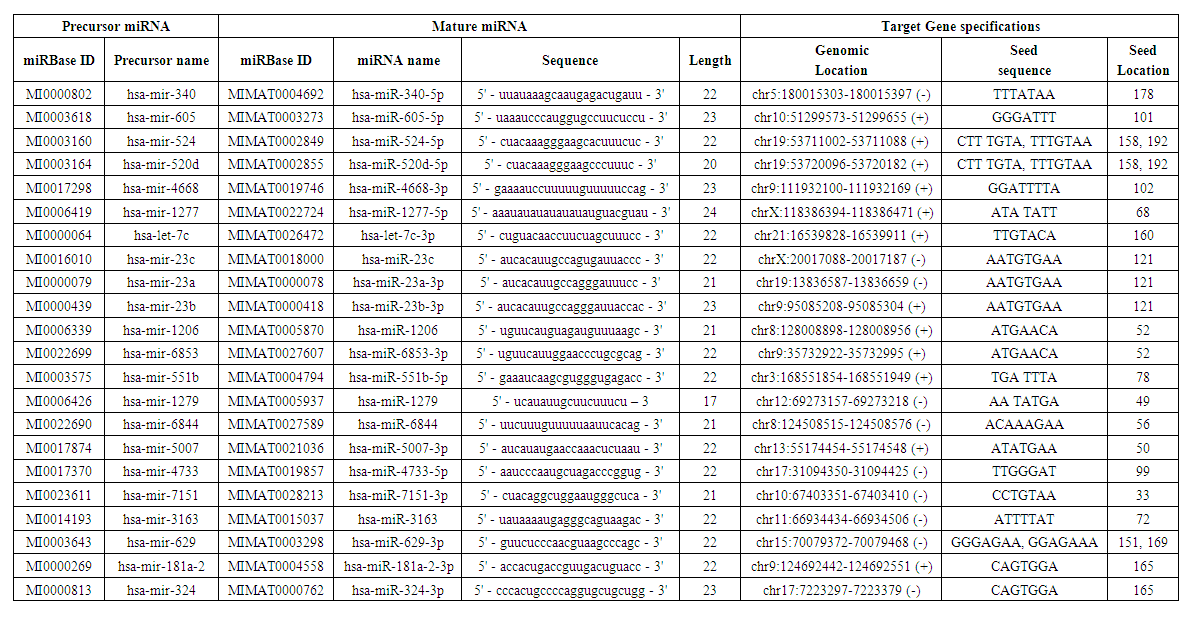
- To explore the miRNA pool targeting LRP5, we use miRDB (MicroRNA target prediction and functional study database http://mirdb.org/). miRDB is an online computational tool for miRNA target prediction and functional annotations [13]. We have used the search term “LRP5” as “Human” “Gene Symbol” and retrieved a list of 22 miRNAs targeting the query gene. We tabulate the details of each miRNA provided by the software. To find out whether these miRNAs target other genes of the predicted network of LRP5, we put each miRNA name as the search item and got the list of all the targets of the query miRNA. From the list we find out the names of the genes associated with LRP5 in its predicted network.
2.3. Confirmation of the miRNA – LRP5 Interaction
- The output of miRDB was confirmed with other three software viz., TargetScanHuman 8.0 (https://www.targetscan.org/vert_80/); RegenDbase (https://regendbase.org/mirna-targets/search) and mirTargetLink 2.0 (https://ccb-compute.cs.uni-saarland.de/mirtargetlink2/).TargetScanHuman predicts biological targets of miRNAs by searching the presence of conserved 8mer, 7mer, and 6mer sites that match the seed region of each miRNA [14]. RegenDbase, the Regeneration Database provides comprehensive set of miRNA-target predictions by integrating mRNA and microRNA expression data from diverse models of regeneration together with data from embryonic and induced pluripotent stem cells [15]. In the form of interactive network visualization, mirTargetLink 2.0 offers information on microRNA-mRNA interactions [16].In case of TargetScanHuman, “Human” is selected as the species and “LRP5” as the query gene symbol. We submit the names of each of 22 miRNAs to find out the interaction of the miRNA and LRP5 transcript. Here not only we got the confirmation of miRNA targeting LRP5 but also the region of their interaction.For RegenDbase, Homo sapiens is selected as “organism” and miRBase mature miRNA accession IDs are used for the final output. It confirms whether a particular miRNA targets LRP5 or not. Each of the mature miRNA names are used as input in mirTargetLink 2.0 to further confirm LRP5 as the target output.
2.4. Preparing a Venn Diagram to Show the Proportion of miRNAs Confirmed by Various Software to Regulate LRP5
- To represent graphically the data retrieved from the entire four target predicting software tools, Venny 2.1 is used. Venny 2.1 (https://bioinfogp.cnb.csic.es/tools/venny/, accessed on 12 September 2023) is a Web-Tool for Creating Venn Diagrams [17]. The list of miRNAs confirmed by each software is used as the input and the output is a Venn diagram showing the proportion of miRNAs confirmed by various software to regulate LRP5.
2.5. Meta-Analysis of MicroRNA Functions Experimentally Proved to Regulate LRP5
- A meta-analysis has been done to find out the functional role of microRNAs in regulation of LRP5. For mining the related experimental data, miRBase (https://www.mirbase.org/) database is used primarily. National Center for Biotechnology Information’s (NCBI) PubMed (https://pubmed.ncbi.nlm.nih.gov/about/) has also been utilized for these experimental resource supports.
3. Result
3.1. Genetic Network of LRP5
- GeneMania identified that LRP5 interacts with 20 others genes viz. SOST, CAPRIN2, DKK1, APCDD1, FZD10, DKK4, AXIN1, WNT7B, FZD4, MESD, FZD1, WNT1, RGL3, RGL2, DKK2, LRP6, LRP5L, WNT5A, FZD8, RGL1 according to Physical Interactions, Co-expression, Predicted, Co-localization, Genetic Interactions, Pathway, Shared protein domains etc. (Figure 1, Table 1).
|
 | Figure 1. LRP5 genetic network showing LRP5 interactions with other related genes as predicted by online software tool GeneMANIA (Adapted from Sengupta & Das, 2023) |
3.2. miRNAs Targeting LRP5 and Its Interacting Partners
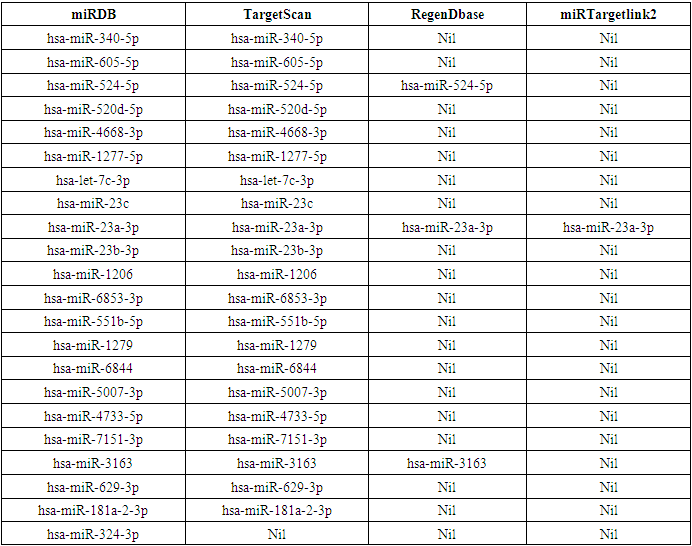
- Data of 22 specific miRNAs targeting LRP5 were retrieved using miRDB database (Table 2). All the 22 miRNAs bind to the 214 bp sequence of 3' UTR region of LRP5. Interestingly several of these miRNAs also target some of the major genes interacting with LRP5 (Table 3).
 | Table 2. List of miRNAs prdicted by online tool miRDB to target LRP5 transcript |
|
3.3. Validation of miRNA-LRP5 Interaction by Other Software
- In this study, we collect responsible miRNAs from miRDB database individually for LRP5. After that, common responsible miRNAs were collected by using Venny online tools. The miRNAs specified by miRDB regulating LRP5 were reconfirmed by other software. Among 22 derivatives of miRDB, TargetScan validated 21, RegenDbase confirmed 3, MirTargetLink 2.0 confirmed only one miRNA targeting LRP5 (Table 4). These proportions of miRNAs confirmed by different software are graphically represented by a Venn diagram (Figure 2). It shows that 22 miRNAs predicted by miRDB to be 100%. Accordingly, targetScan confirms 95.4%, RegenDbase confirms 13.6%, MirTargetLink 2.0 confirms 4.5% of total miRNAs predicted by miRDB targeting LRP5. All the four software confirm only one (4.5%) miRNA viz. hsa-miR-23a-3p. Three software i.e., miRDB, TargetScan and RegenDbase commonly confirm two miRNAs (9.1%) viz hsa-miR-524-5p and hsa-miR-3163. Two software i.e., miRDB and TargetScan confirm eighteen miRNAs (81.8%) viz. hsa-miR-340-5p, hsa-miR-605-5p, hsa-miR-520d-5p, hsa-miR-4668-3p, hsa-miR-1277-5p, hsa-let-7c-3p, hsa-miR-23c, hsa-miR-23b-3p, hsa-miR-1206, hsa-miR-6853-3p, hsa-miR-551b-5p, hsa-miR-1279, hsa-miR-6844, hsa-miR-5007-3p, hsa-miR-4733-5p, hsa-miR-7151-3p, hsa-miR-629-3p, hsa-miR-181a-2-3p. One miRNA viz.hsa-miR-324-3p is confirmed only by miRDB.
|
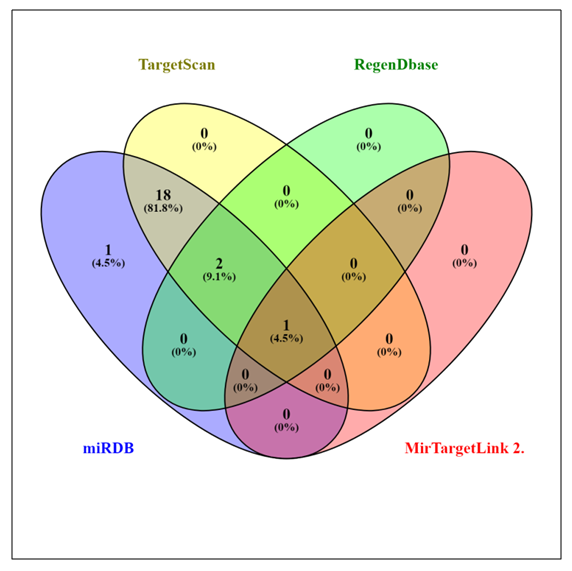 | Figure 2. Venn diagrams created by using Venny 2.1 graphically representing the data retrieved from the entire four target predicting software tools |
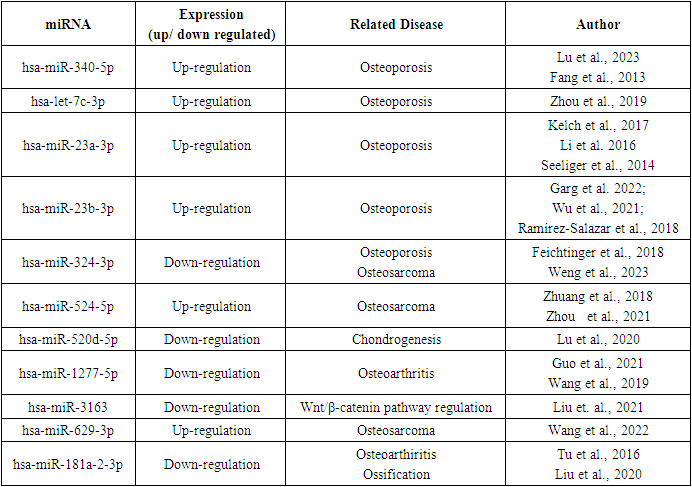
3.4. Functional Relevance of the miRNAs Targeting LRP5
- A total of 20 studies were identified in the systematic review indicating important roles of the miRNAs enlisted in this work in osteoporosis, low BMD conditions and other bone related diseases. These studies depicted an altered expression level of 10 miRNAs from our list (Table 5) in bone related abnormalities. One miRNA hsa-miR-3163 has been reported [41] to regulate oncogenic Wnt/β-catenin pathway which is also highly associated with bone development.
|
4. Discussion
- The role of LRP5 in bone metabolism is well established through scientific studies. Mutations, low expression, and many other variations in this gene are correlated with the development of bone related diseases [12]. Eukaryotic genes are regulated post transcriptionally by miRNAs [42]. The regulation of any gene by miRNAs is resulted through a complex regulatory network involving many other genes and factors [43].Like other genes, LRP5 also functions through a network of associated genes. We have identified twenty genes interacting with LRP5 by various means. Interestingly, many of the miRNAs targeting LRP5 also target its (LRP5) interacting partners. This supports the idea of the existence of a miRNA-LRP5 regulatory network yet to be explored by scientific community.The list of miRNAs provided by miRDB is validated with other software also. The very particular hsa-miR-23a-3p confirmed by all the software has been reported to be up-regulated in both male and female osteoporotic patients [44]. Over expression of microRNA-23a targeting LRP5 prevents osteogenic differentiation in human bone marrow-derived mesenchymal stem cells (hBMSCs) and down regulation of the same miRNA enhanced the process of osteogenic differentiation of hBMSCs [45]. This miRNA has also been reported to be a diagnostic and prognostic marker of osteopororsis [46]. Elevated serum level of miR-340-5p is observed osteoporotic postmenopausal women. Circulating miR-340-5p has been proposed as an osteo-miRNAs in postmenopausal women and potential biomarker of osteoporosis in postmenopausal women [47]. It has also been found to prevent Wnt/β-Catenin signaling pathway by decreasing β-catenin protein levels by 2- to 5-fold [48]. In 2019, Zhou et al. [49] reported that upregulated miRNA let-7c inhibits Wnt/β-catenin signaling and in postmenopausal osteoporotic patients. Up-regulated miR-23b-3p is associated with osteoporotic hip fractures and suggested as the risk factor for osteoporosis by [50]. It has also been proposed as the diagnostic marker for human osteoporosis and fragility fracture [51]. High serum level of miR-23b-3p thus may be a key regulator of bone mineral density [52]. Interestingly down-regulation of miR-324 is correlated with osteoporortic fractures. The serum level of miR-324 is positively correlated to bone mineral density [53]. MicroRNA-324-3p is found to inhibit osteosarcoma (OS) progression. Reduced expression is identified in in OS cell lines and tissues [54]. A role of circITCH/miR-524/RASSF6 axis is reported to suppress OS progression [55]. Another report of significant up-regulation of miR-524 in OS denotes its role in promoting cell proliferation in the disease [56]. The up-regulation and down-regulation of miR-520d-5p have been reported to promote and inhibit chondrogenesis respectively, and regulate chondrocyte metabolism [57]. Down regulation of miR-1277-5p is associated with osteoarthritis development [58,59]. A role of miR-3163 is proved in activating Wnt/β-catenin pathway in pancreatic cancer. But the functional role has not yet been investigated in Wnt/β-catenin pathway related to bone metabolism. Anti-calcification effects of hsa-miR-629-3p are reported in osteogenic differentiation-induced human aortic valve interstitial cells (hVICs) [60]. A role of miR181 family members has been depicted in chondrocyte differentiation and formation [61]. It is reported that MicroRNA-181 regulates the development of Ossification of Posterior longitudinal ligament [62].
5. Conclusions
- It can be concluded that there is functional significance of predicted miRNA in regulation of LRP5. These regulatory miRNAs may influence the expression level of LRP5, a key regulator of bone mineral density and its interacting partners. This altered expression in LRP5 network may hinder the down signaling cascade of WNT-β catenin pathway. This ultimately may affect the bone metabolism and develop into osteopororsis. There are reports of regulatory functions of about 11 miRNAs from our list. The rest 11 need more concern of the scientific world for finding out their plausible role(s) in bone metabolism and osteoporosis consequently.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML