-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Bioinformatics Research
p-ISSN: 2167-6992 e-ISSN: 2167-6976
2019; 9(2): 56-67
doi:10.5923/j.bioinformatics.20190902.02

In-Silico Analysis of Three Transcription Factors Contributing to Repair and Regeneration of Lung Epithelial Progenitor Cells
Mohamed L. Salem1, 2, Ahmed M. Azlohairy3, Ismail Atia2, 4, Heba Wassfy2, 5, Abdel-Aziz A. Zidan2, 6, Gábor Gyulai7
1Department of Zoology, Faculty of Science, Tanta University, Tanta, Egypt
2Center of Excellence in Cancer Research, Medical Campus, Tanta University Teaching Hospital, Tanta, Egypt
3Genetics Department, Faculty of Agriculture, Zagazig University, Egypt
4Zoology Department, Faculty of Science, Alazhar University, Assuite, Egypt
5Faculty of Engineering, Tanta University, Tanta, Egypt
6Faculty of Science Damanhur University, Damanhur, Egypt
7Institute of PhD Schools, St. Stephanus University, Gödöllö, H, Hungary
Correspondence to: Mohamed L. Salem, Department of Zoology, Faculty of Science, Tanta University, Tanta, Egypt.
| Email: |  |
Copyright © 2019 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Cell differentiation processes of stem cells have unique pathways controlled by certain genes including EYA1 (Eyes Absent), SIX1 (Sine oculis homeobox), and SOX9 (Sry-related high mobility group BOX). It was reported that these genes act as transcriptional factors (TFs) for the repair and regeneration of the progenitor cells as well as of damaged tissues, however the function of these genes are not fully characterized and understood. Different bioinformatics tools were used in this study to analyze four transcriptional factors (TFs of EYA1, SIX1 and SOX9) in human and compare them with their orthologs in eleven different organisms by tools to provide more knowledge about their functions in stem cells. The present investigations showed that SOX9 was neither present in Ceanorhabditis elegans (nematode; roundworm) nor in Cavia porcellus (domestic guinea pig). We applied multiple sequence alignments (MSA) for selection isoforms of SOX9 protein showed conserved domains among different species. EYA1 protein showed wide range of alternative isoforms in Homo sapiens. High levels of SIX1 were detected in all human body tissues. Gene expressions were investigated to determine the expressions of genes in body tissues. The interaction between TFs proteins was confirmed by gene interaction network (GIN) which showed close relations between EYA1 and SIX1. The results of this study shed a light on the differential gene expression level of studied genes in different tissues damaged by cancer and their different repair mechanisms. The presence or absence of these TF genes may be an indicator of the developmental differentiation between invertebrates and vertebrates.
Keywords: Bioinformatics' analysis, Progenitor cells, Transcription factors, Lung stem cells
Cite this paper: Mohamed L. Salem, Ahmed M. Azlohairy, Ismail Atia, Heba Wassfy, Abdel-Aziz A. Zidan, Gábor Gyulai, In-Silico Analysis of Three Transcription Factors Contributing to Repair and Regeneration of Lung Epithelial Progenitor Cells, American Journal of Bioinformatics Research, Vol. 9 No. 2, 2019, pp. 56-67. doi: 10.5923/j.bioinformatics.20190902.02.
Article Outline
1. Introduction
- Stem cells are defined as undifferentiated cells which have the potential for self-renewal and develop into various cell types. In particular, lung stem cells are capable of carrying out different functions because of their ability of self-renewal and pluripotency. The lung has a much slower turnover of putative lung progenitor cells, unlike other epithelial tissues that undergo rapid regeneration [1,2]. Transcription factors plays an essential roles in regulation of gene expression [3]. TF genes of EYA1, SIX1, SOX9 have recently attracted the attention due to their multiple roles as transcriptional factors (TF) in controlling the transcription of DNA to mRNA by promoting or blocking the recruitment of RNA polymerase to specific genes [4]. EYA1-4 and sine oculis family genes (SO; which is a member of the SIX family of homeobox transcription factors, are unable to initiate eye development in non-retinal tissues) [5] exhibited interactions to regulate the development of many organs [6-9]. The eyes absent proteins act as transcriptional co-activators [10]. EYA1 switches SIX1 function from suppression to activation condition in the nucleus, resulting in transcriptional activation through recruitment of co-activators regulating precursor cell proliferation and survival during organogenesis [11]. EYA1 and SIX1 are important transcription factors for the lung epithelial stem/progenitor cells maintenance. Mice, deficient in EYA1 or SIX1, do not have the epithelial progenitor cell markers; they highly express differentiation markers in their lungs. In addition, their lungs are markedly hypo plastic with decreased epithelial branching capacity and intensive mesenchymal cellularity [12,13]. SIX1 are responsible for developing skeletal muscles and supports gene expressions related to functioning of developing progenitor cells. SIX1 is a critical TF in skeletal muscle development, and also in eye and kidney development. Numerous studies have demonstrated that SIX gene family play an important role in organogenesis [11,14-16]. Moreover, SIX1 is able to drive the transformation of slow-twitch fibers to the fast-twitch phenotype in synergy with its cofactor EYA1 in muscles of adult mice [5]. SOX9 is a member of high mobility group-box class DNA binding protein family of transcription factors, it is required for branching morphogenesis in the lungs and for controlling alveolar epithelial progenitor cell proliferation [17,18]. Upregulation of SOX9 expression was reported to increase cell proliferation [19-21] associated with a poor lung ADC (Antibody-Drug Conjugate) survival [1]. However, the functional role of SOX9 in lung cancer has not been fully elucidated. Furthermore, the upstream oncogenic molecular pathways regulating SOX9 overexpression in lung ADC have not been completely delineated.SOX9 is highly expressed in the distal epithelial progenitors in embryonic lung, nonetheless conditional deletion of SOX9 does not affect progenitor cell behavior in lung [17,22,23].In this study, genes and proteins sequences of identified EYA1, SIX1, SOX9 transcriptional factors in twelve different organisms were analyzed. Constructed Phylogenetic trees of TFs protein sequences showed their cladistics relation based on their structure and function. Moreover, TFs protein domains, alternative splicing [24], expression patterns, and interaction network were analyzed to elucidate these TFs genes in human tissues. This investigations shwed the importance of these genes in stem cell differentiation and their relations in many syndromes and functional disorders. Those genes were chosen to provide more knowledge about their role not only in human but also in other living organisms to investigate their functional roles. The results of this investigation may open a new venue to repair lung defects due to cancer or other disorders.
2. Material and Methods
- Data resourcesThe EYA1, SIX1, and SOX9 TF protein sequences used for analysis and phylogenetic tree construction were obtained from the Uniprot database (). Transcript data used for analyzing the alternative splicing and the functional protein domains were obtained from NCBI database (http://www.ncbi.nlm.nih.gov). Expression analysis datasets were obtained from BioGPS database (http://Biogps.org/#goto).Protein interaction networks were obtained from Gene Card human gene database (http://www.genecards.org/). Identification of protein sequences and phylogenetic tree constructionEYA1, SIX1 and SOX9 TF protein sequences were obtained from the Uniprot database (http://www.uniprot.org//) and used as templates from twelve different spices To understand how different evolutionary changes of the structure did not affect conservative function of Homo sapiens (Human), Pan troglodytes (common chimpanzee), Macaca mulatta (Rhesus monkey), Gorilla gorilla (western gorilla), Mus musculus (mouse), Rattus norvegicus (Norway rats), Cavia porcellus (domestic guinea pig), Canis lupus (gray wolf); bird Gallus gallus (red junglefowl); frog Xenopus laevis (African clawed frog); fish Danio rerio (zebrafish); and roundworm Caenorhabditis elegans (nematode). Data were downloaded, analyzed, and phylogenetic trees were constructed using MEGA6 by the neighbor-joining method.Protein Domains analysis and identification of alternative splicingThe functional TF protein domains of EYA1, SIX1 and SOX9 were obtained from NCBI database and analyzed by using the Pfam sequence tool (protein family database which contains information about protein domains and families) (https://www.ncbi.nlm.nih.gov/Structure/cdd). Alternative splicing of each gene was obtained from NCBI database. Expression analysis in human body tissues The gene expression analysis for TF EYA1, SIX1and SOX9 was obtained from BioGPS database (http://Biogps.org/#goto) representing samples of Human tissues at normal phase. More than 76 samples of tissues were measured.Protein interaction network construction Protein interaction network were obtained from Gene Card (Human gene database) (http://www.genecards.org/), and were illustrated the interacting proteins searched for. This search was based on published papers experimentally proven and accepted.
3. Results
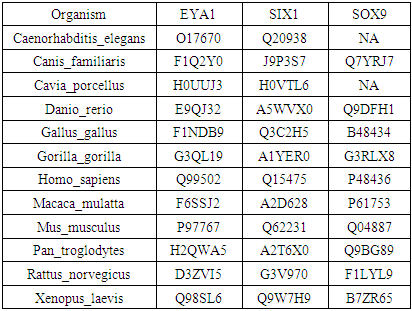
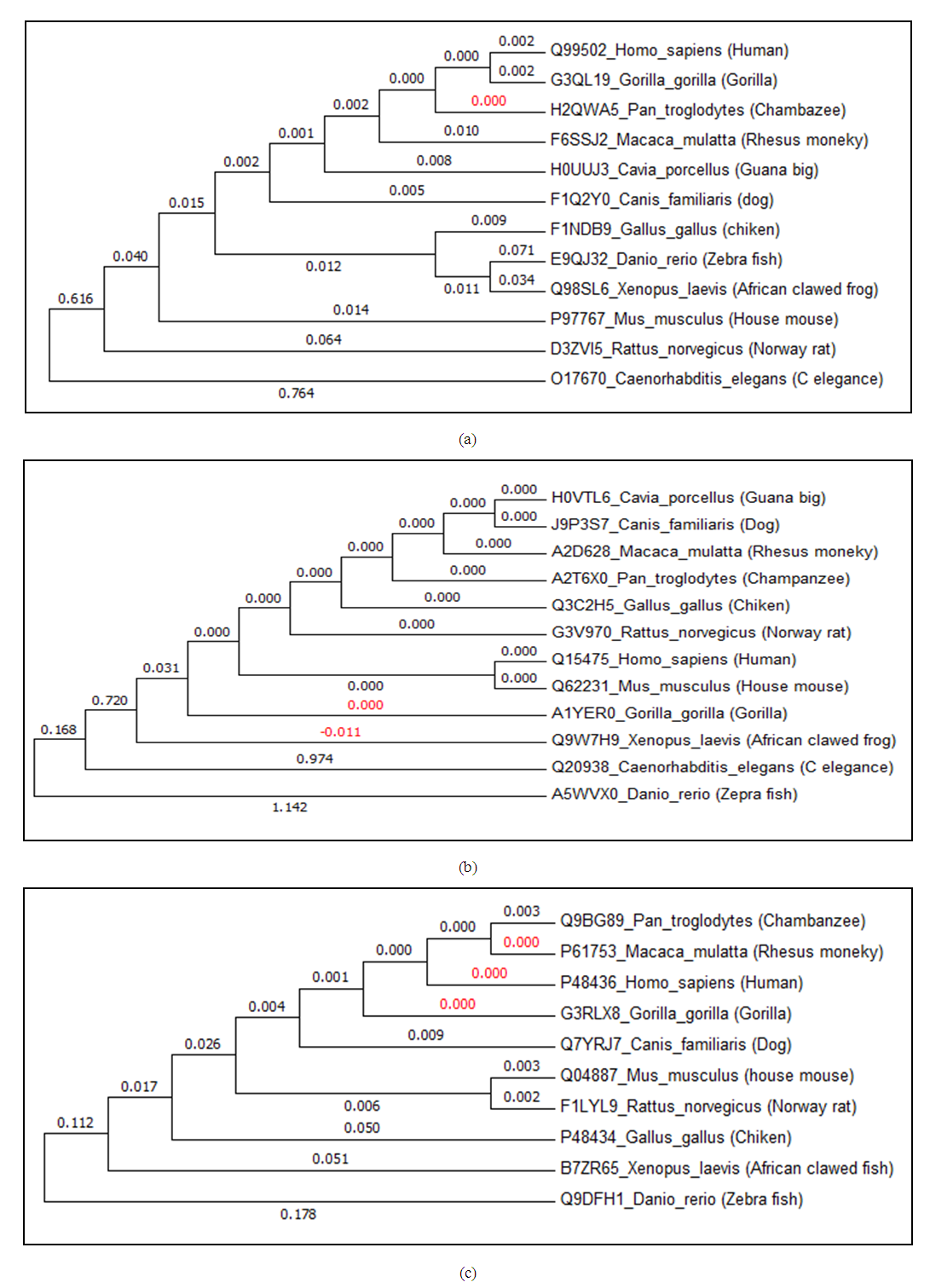
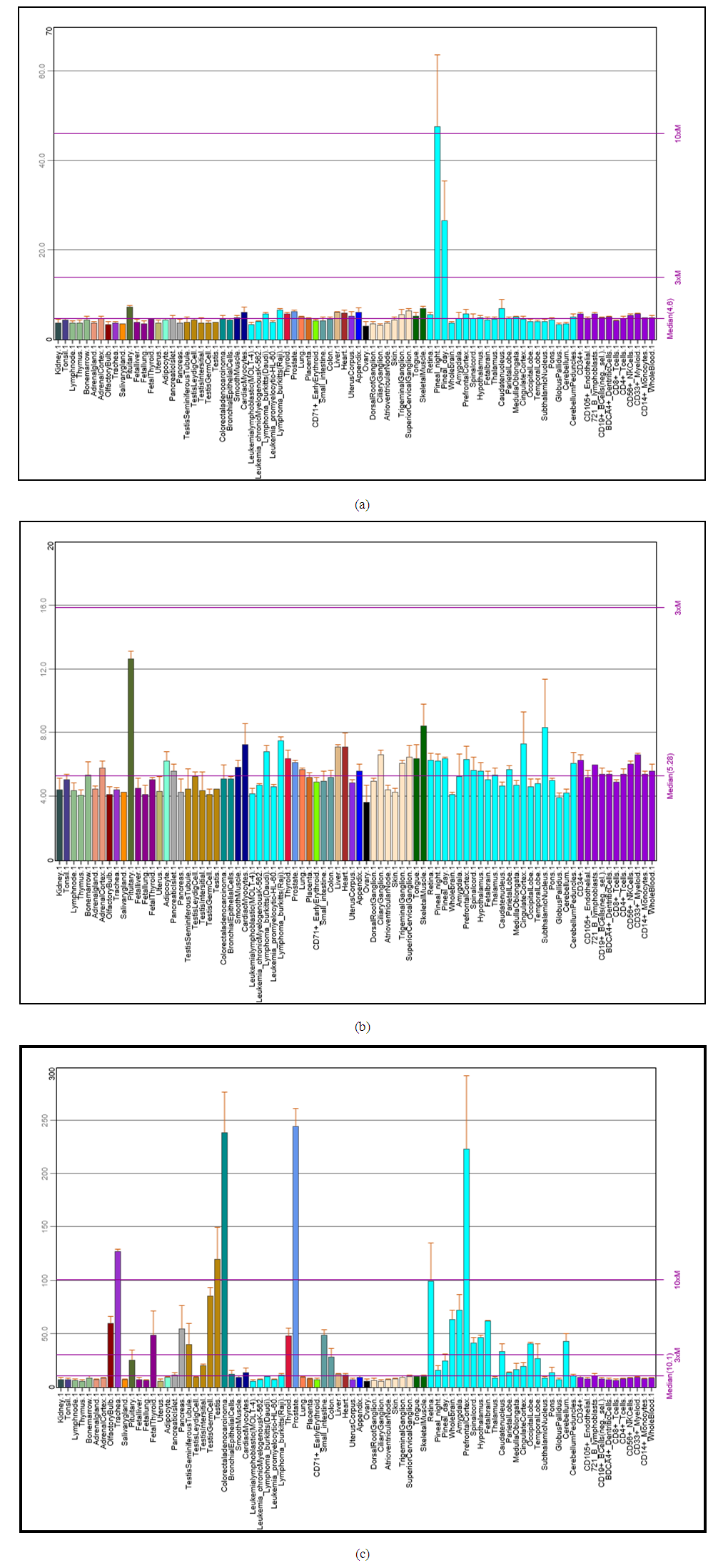
- Identification of TF proteins in species and phylogenetic tree constructionDifferent methods were used with all sequences to generate a multiple sequence alignment using [25,26]. In order to analyze TF gene sequences, we used MEGA6 program [27] to further analyze the function and conservation levels of these transcription factors, and to explain the evaluation between species. By using Neighbor-joining (NJ) method we constructed the phylogenetic tree of these genes. We assumed that the three TF exist in all species, however, in contrast to the expectation, SOX9 was not found in Caenorhabditis elegans and Cavia porcellus.The same results were obtained using Phylip [28]. This observation indicated that the function of the SOX9 gene in this species may be regulated by other genes, or may be lost during differentiation due to their live cycle. The role of gene as sex determinate not as transcriptional factor may indicate developmental differences between vertebrates and invertebrates (Table 1).
|
|
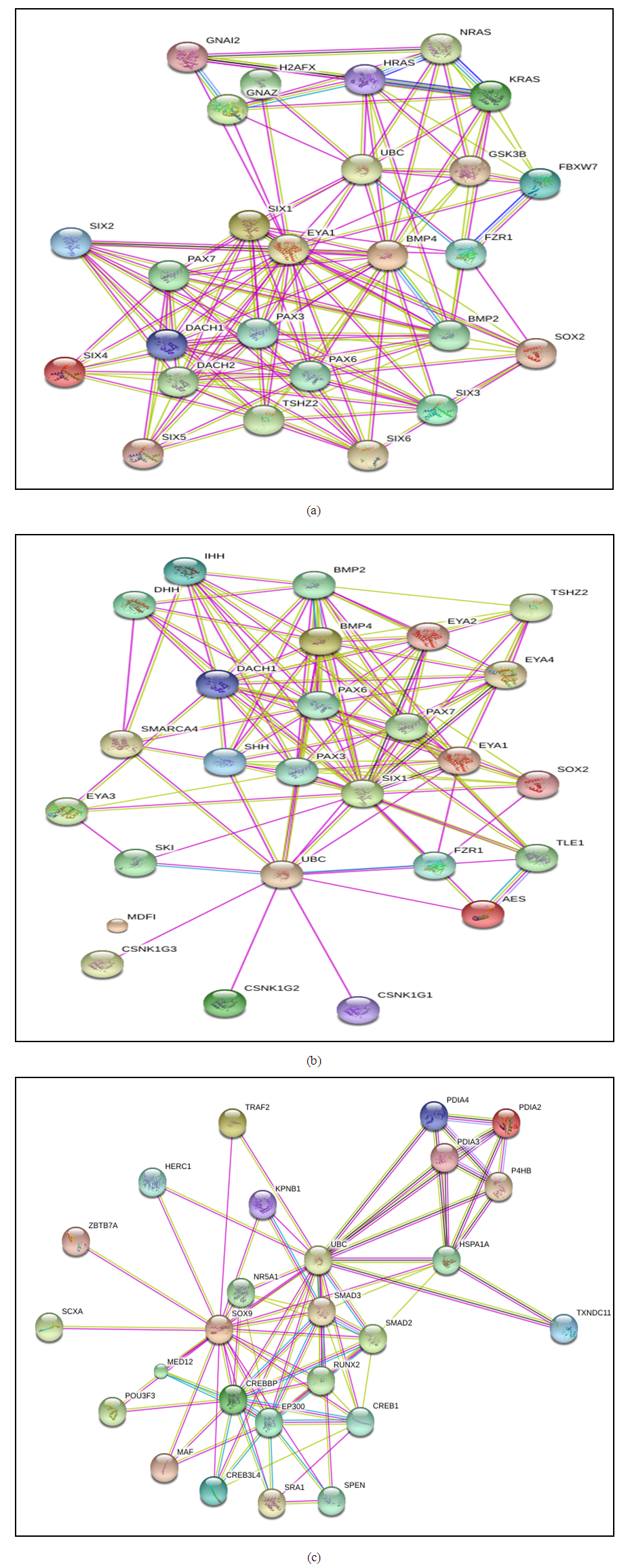 | Figure 4. The TF proteins interaction networks of (A) EYA1, (B) SIX1, and (C) SOX9. Data were obtained from Gene Card (human gene database) and illustrated the interacting proteins |
4. Discussion
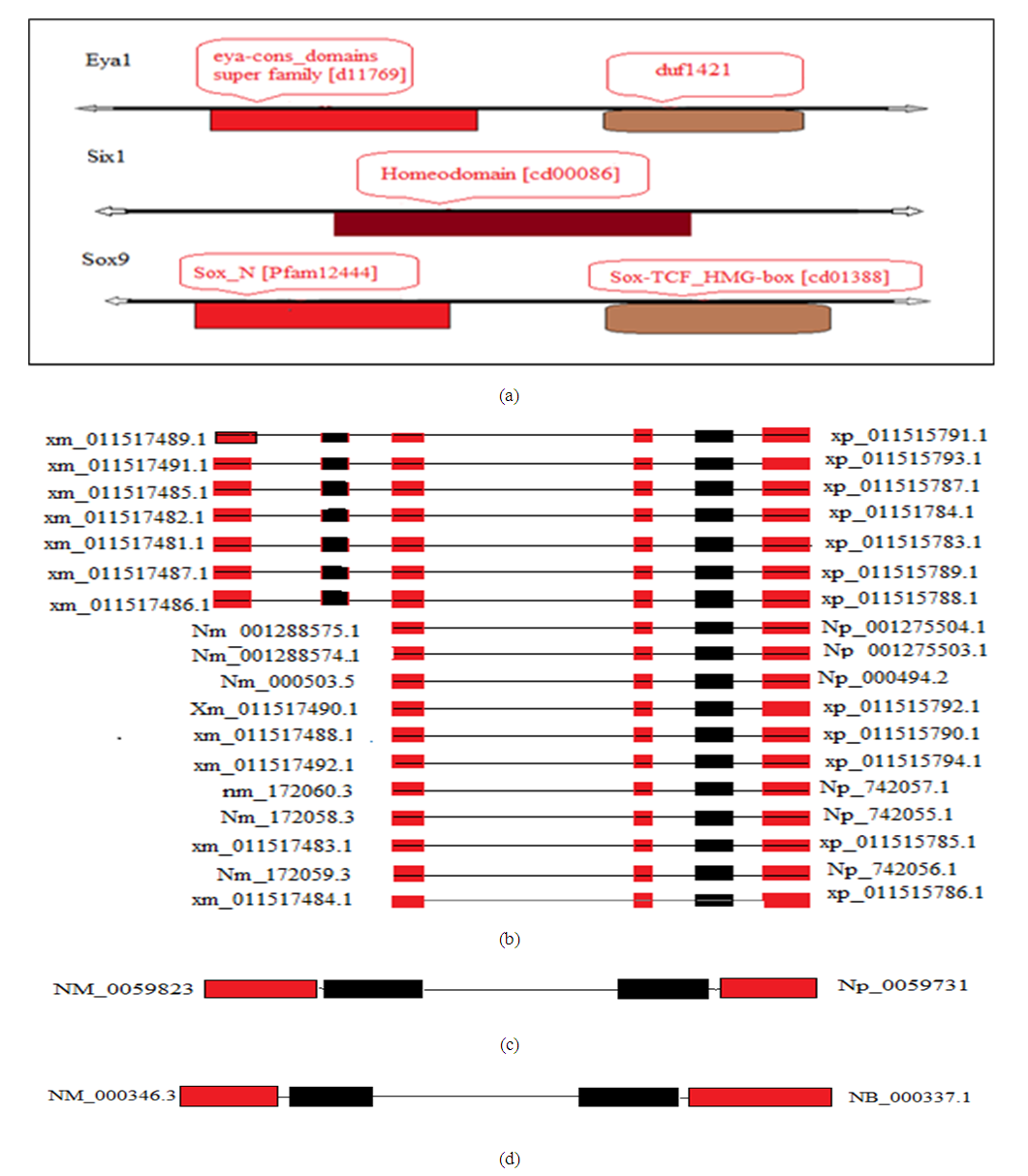
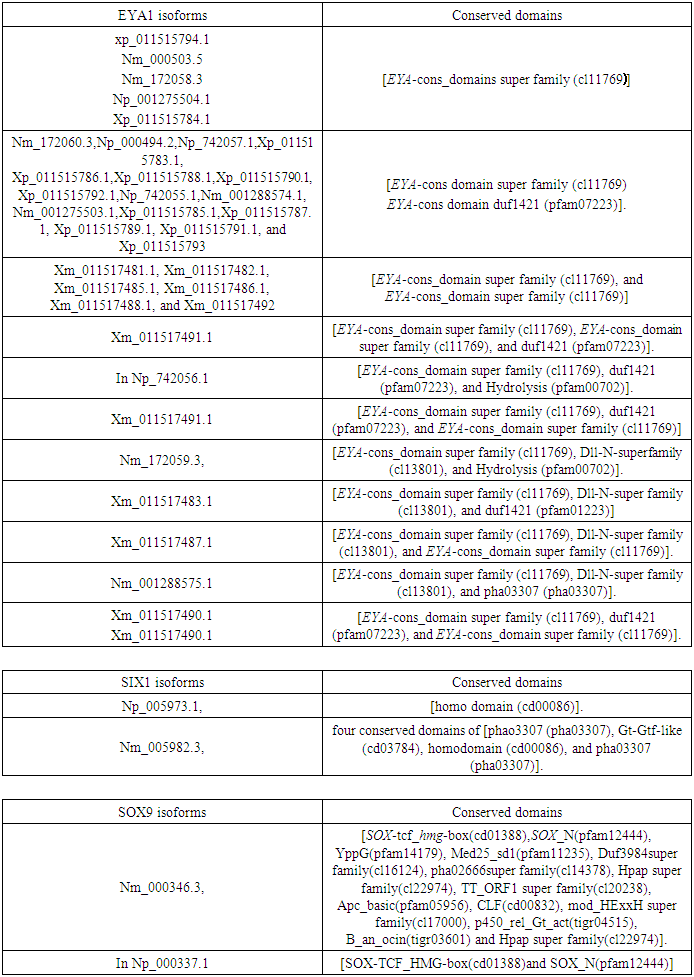
- The phylogenetic trees mainly followed the evolutionary relationships among the species. We found that SOX9 doesn’t exist in Cavia porcellus, Caenorhabditis elegans, which may indicate that the function of SOX9 is regulated by other TFs in these organisms. Conserved domains of EYA1, SIX1, and SOX9 were found. EYA1 showed more than 36 alternative isoforms, which indicated that alternative splicing is a very important source of protein diversity, and accounts for the majority of different protein functions [31,32]. Expression analysis of genes in human body tissue showed normal levels detected in EYA1, and SOX9. However, high levels of expressions were detected in sex organs. High levels of SIX1 expression were detected in all body tissues, which indicate that SIX1 is mainly responsible for all organ development in order to its somatic function. These results support the relation with function of developing progenitor cells associated with tumor progression and metastasis, which provides a mechanism for activation of specific gene targets, including those regulating precursor cell proliferation during organogenesis [11,14,35,36]. Interaction network of the three TF genes showed strong interactions, and confirmed that EYA1 and SIX1 interact in a complex way, and SIX1 interacts with EYA1 in the development of several organs in the human body.
5. Conclusions
- The three transcription factors were found to have crucial roles in the development and function of organs in animal bodies. Focusing to lung, our results may provide new knowledge about these TF genes and open new prospects to further research which can help to understand repairs of lung defects in order to cancer or other diseases.
ACKNOWLEDGEMENTS
- This work has been supported by a grant (ID# 5245) funded from the Science and Technology Development Fund (STDF), Ministry of Scientific Research, Egypt to prof. Mohamed L. Salem, the Principal investigator of this project.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML