-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Bioinformatics Research
p-ISSN: 2167-6992 e-ISSN: 2167-6976
2019; 9(2): 45-55
doi:10.5923/j.bioinformatics.20190902.01

Bioinformatics Analysis and the Revelation of Thirteen Novel Mutations in Human LH-B Gene Related to PCOS
Nidal Essa1, Abdelrahman H. Abdelmoneiom2, Razan M. Badawi2, Afnan Mohamed Mohamed Khair2, Rania Anwer2, Amel Nasir Eltayeb Ali2, Tebyan Ameer Abdelhameed Abbas2, Mohamed A. Hassan2, 3
1Faculty of Medical Laboratory Sciences, University of Medical Sciences and Technology, Khartoum, Sudan
2Department of Bioinformatics, Africa City of Technology, Khartoum, Sudan
3Department of Bioinformatics, DETAGEN Genetic Diagnostics Center, Kayseri, Turkey
Correspondence to: Nidal Essa, Faculty of Medical Laboratory Sciences, University of Medical Sciences and Technology, Khartoum, Sudan.
| Email: |  |
Copyright © 2019 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Luteinizing hormone beta subunit (LH-B) (protein ID P01229) is gonadotropin hormone secreted from the anterior pituitary belongs to the glycoprotein family, mapped on chr19p13.3 and consists of three exons, three transcript variants with two phenotypes encoded for one protein known as Lutropin subunit beta (P01229) It has a central role in promoting spermatogenesis and ovulation by stimulating the testes and ovaries for steroidogenesis. PCOS is a common endocrinopathy affecting of women within the reproductive age, abnormal ovulation associated with a high level of LH as a result of gene polymorphisms lead to infertility problems. In this study, we used various computational approaches to identify nsSNPs which probably be deleterious to the structure and/or function of LH-B protein that might be associated with polycystic ovary syndrome. The data on human LH-B gene was retrieved from dbSNP/NCBI. Eleven different bioinformatics prediction algorithms; SIFT, Polyphen, PROVEAN, SNAP2, Pmut, PhD-SNP, I-Mutant and Project Hope were used to analyze the effect of nsSNPs on functions and structure of the LH-B protein, and RaptorX for protein modeling and Chimera for visualization of the model, in addition, we used PolymiRTS to detect SNPs on miRNA binding sites. After retrieval of SNPs from the NCBI database, 140SNPs were classified as missense SNPs. From functional analysis software, 39 SNP were predicted to be deleterious then they analyzed by disease-related software 13 SNPs, when checked for protein stability, 12 of them decreased protein stability and one SNP increased its stability. Hence, application of these 13 novel mutations through genetic studies will contribute towards our understanding of the pathophysiology of PCOS.
Keywords: PCOS, In Silico analysis, LH-B, Folliculogenesis, Computational analysis, SNPs, Mutation
Cite this paper: Nidal Essa, Abdelrahman H. Abdelmoneiom, Razan M. Badawi, Afnan Mohamed Mohamed Khair, Rania Anwer, Amel Nasir Eltayeb Ali, Tebyan Ameer Abdelhameed Abbas, Mohamed A. Hassan, Bioinformatics Analysis and the Revelation of Thirteen Novel Mutations in Human LH-B Gene Related to PCOS, American Journal of Bioinformatics Research, Vol. 9 No. 2, 2019, pp. 45-55. doi: 10.5923/j.bioinformatics.20190902.01.
Article Outline
1. Introduction
- Luteinizing hormone beta subunit (LH-B, OMIM 152780) as hypothalamus-pituitary-gonadal axis (HPO) gene, responsible for the production of steroid hormones and ovulatory process, mainly secreted from the anterior pituitary gland, and it belongs to glycoprotein family which has two subunits; an identical alpha subunit and a hormone-specific beta subunit [1,2]. LH-B also known as Lutropin beta chain, mapped onchr19p13.3 (1111bp, linear DNA) and consists of three exons, three transcript variants with two phenotypes encoded for one protein known as Lutropin subunit beta (P01229)which consists of 141 amino acids and 15,345(Da). It has a central role in promoting spermatogenesis and ovulation by stimulating the testes and ovaries for steroidogenesis [3], during ovulation, which is mainly LH dependent process, requires surge of LH-B levels to initiate the steroidogenesis which is essential for follicular development and corpus luteum formation through binding of LH to its high affinity receptor, LH/choriogonadotropin receptor (LHCGR) in specific theca and granulosa cells for androgen production mainly progesterone and estrogen, which maintain the endometrium for conception [4,5]. If conception or implantation fails to occur, the endometrium is shed and the corpus luteum atrophies to an atretic follicle. Therefore, anovulation due to abnormal LH signaling leads to a liberal role in amplifying ovarian androgen production that might be associated with polycystic ovary syndrome (PCOS) [5-7].The appearance of PCOS as Stein & Leventhal syndrome [8] in 1935 as first report of the disease also described it as a syndrome of contradictory, because there is heterogenecity of both ovarian morphology and clinical findings such as hirsutism, obesity, infertility and polycystic ovary, also this heterogenecity extended to its genetic background with polygenic mode of inheritance. In addition to other genetics studies revealed that genes responsible from signal transduction pathways which control many of them such as steroidogenesis, steroid hormones action, gonadotropin action and regulation, insulin action and secretion, energy homeostasis and chronic inflammation show abnormal expressions among PCOS population [9-12]. Hyperandrogenaemia as well as anovulation is the central hallmarks of PCOS, the majority of females with PCOS suffering from abnormal menstrual cycles and anovulatory problem. Many studies detected abnormal elevation of LHB due to gene polymorphisms which resulted in bioactivity changes in structure and even through the functional part of the hormone associated with anovulation among PCOS women. But until now, the relationship between LH signaling pathway and PCOS has not been defined clearly. Therefore a few previous studies concerning the role of LHb and LHCGR polymorphisms in PCOS have provided conflicting results such as [13-15]. Moreover, LHCGR polymorphisms may also lead to biological activity changes that support the worse effect of abnormal LHB action because of its transmembrane integral protein acts as a G-protein-coupled receptor superfamily controlling the LHB level in the theca cell membrane [5-18].It has been reported that PCOS women of different ethnicity display different clinical manifestations. Amal Salah et al revealed that LHβ G1052A and LHCGR G935A genes polymorphisms are associated with increased risk of PCOS in Egyptian women, especially in obese cases because obesity aggravates menstrual irregularity and increases serum total testosterone level. Thus, Hyperandrogenism can affect the follicle growth and metabolic process, playing a key role in the occurrence and the development of PCOS in the obese women [19,20]. Additionally, two missense point mutations were discovered among PCOS women in LHβ gene (Trp 8 Arg and Ile 15 Thr) with obesity association only in Japanese population, while obese PCOS women from north European countries showed lower frequency [5]. Also, another variation that occurs due to ethnicity factor reported that LHβ gene variant G102A in exon 3 (Gly102Ser) was found to be higher in Singapore Chinese women who had menstrual disorders in contradiction to Korean research that found no difference in elevated blood glucose associated with insulin resistance as a metabolic feature of PCOS also associated with LHb 1052A allele [24-26]. High level of blood glucose associated with insulin resistance as a metabolic feature of PCOS also associated with LHB 1052A allele. While LHB 1052A allele carriers showed lower LH level and higher total and free testosterone levels due to SNPs effect on gene bioactivity.The aim of computational biology is to permit researchers to perform experiments in silico on digital virtual models of biological molecules such as cells, organs and organisms. These virtual models hold great promise for enhancing the pace and covering the scope of biomedical research by freeing scientists from biological experimental barriers associated with ethical restrictions [27-30].This leading and unique study benefits from the in silico analysis tools to build up knowledge about PCOS and to determine the influence of various SNPs in LH-B gene using in silico prediction software and assessing their effects on the structure/function of LH protein that may have an important role in disease susceptibility. This role will be examined through wet laboratory work as a part of PhD project that will enhance in understanding the genetic mechanisms of the disease through these useful tools. We followed the same protocol that was used for FSH-B gene analysis [31].
2. Material and Methods
- 2.1. Data retrieval: The data on human LH-B gene was collected from the National Center for Biological Information (NCBI) web site. The SNP information (protein accession number and SNP ID) of the LH gene was retrieved from the NCBI dbSNP (http://www.ncbi.nlm.nih.gov/snp/) [1] and the protein sequence was collected from Uni Prot databases [3].2.2. SIFT (Sorting Intolerant from Tolerant): https://sift.bii.a-star.edu.sg/: Is an online tool that predicts if an amino acid substitution affects protein function or not by using sequence homology. It performs analysis based on different algorithms and it interprets the homologous sequences using the Swiss-Prot (version 51.3) and TrEMBL (version 34.3). It gives scores to each amino acid residue ranging from zero to one. The threshold intolerance score for SNPs is 0.05 or less [32].2.3. Polyphen-2 (Polymorphism Phenotypingv2), http://genetics.bwh.harvard.edu/pph2/: Is used to predict the possible impact of an amino acid substitution on both structure and function of protein by analysis of multiple sequence alignment and protein 3D structure. It estimates the position specific independent count score (as probably damaging, possibly damaging or benign according to the score ranging from (0–1). [33].PSIC) for every variant and then determines the difference between them, the higher the PSI, the higher the functional impact of the amino acid on the protein function may be. Prediction outcomes could be classified2.4. Provean (Protein Variation Effect Analysis) (http://provean.jcvi.org/index.php): This is a software tool that predicts the impact of amino acid substitution on the biological function of the protein when we entered the protein FASTA sequence along with amino acid substitutions as input query for analysis. The outcome product according to the threshold score of −2.5 as tolerated or deleterious effect [34].2.5. SNAP2 (Screening of Non-Acceptable Polymorphism 2; https://rostlab.org/services/snap2web/): Is a neural network dependent software tool that distinguishes between effect and neutral variants/non-synonymous SNPs by taking a variety of sequence and variant features into account. After entering the protein FASTA sequence as input query while the output predication as a neutral, non- neutral effect [35].2.6. PhD-SNP (Predictor of human Deleterious Single Nucleotide Polymorphisms; http://snps.biofold.org/phd-snp/phd-snp.html):It is an online Support Vector Machine (SVM) based classifier, which is optimized to predict if a given single point protein mutation can be classified as disease-related or as a neutral polymorphism, also we used the protein FASTA sequence as input query besides the residues change to obtain the output results [36].2.7. SNPs &GO (Single nucleotide polymorphism & Gene Ontology, PHD-SNP (http://snps.biofold.org/snps-and-goinformation evolutionary information and function as encoded in the Gene Ontology terms and other outperforms with available predictive methods [37].2.8. P-mut (http://mmb2.pcb.ub.es:8080/PMut): Is a web-based tool for the annotation of pathological variants on proteins based on the uses of neural networks. The output display according to the pathogenicity index ranged from 0- 1; more than 0.5 scored as single pathological mutations [38].2.9. I–Mutant 3.0 (http://gpcr2.biocomp.unibo.it/cgi/predictors/I-Mutant3.0/I-Mutant3.0.cgi): It is a neural network based tool, predicts the change in the stability of the protein upon mutation The output is obtained in the form of protein stability change upon mutation and Gibbs-free energy change (DDG) either increased or decreased stability [39].2.10. Project Hope (http://www.cmbi.ru.nl/hope/): It is a new online web-server to search protein 3D structures (if available) by collecting structural information from a series of sources, including calculations on the 3D coordinates of the protein, sequence annotations from the Uni Prot database, and predictions by DAS services. Submission of protein sequence and the mutant variants as input information to the web server which displays the output as textual report supported by figures and animations [40].2.11. RaptorX (http://raptorx.uchicago.edu/): Is a web server predicting structure property of a protein sequence without using any templates. It outperforms other servers, especially for proteins without close homologs in PDB or with very sparse sequence profile. The server predicts the tertiary structure [41].2.12. PolymiRTS Database 3.0: As well as it is a database of naturally occurring DNA variations in microRNAs (miRNA) seed region and miRNA target sites, it is used to predict 3un-translated region polymorphism (3’ UTR) effect upon microRNAs and their target sites which may influence miRNA-mRNA interaction, causing impact on miRNA-mediated gene repression. PolymiRTS database was made by examining 3UTRs of mRNAs in human and mouse for SNPs in miRNA target destinations. Then, the impact of polymorphism on gene expression and phenotypes are identified and then connected in the database. The PolymiRTS data base also includes polymorphism in target sites that have been supported by a variety of experimental methods and polymorphism in miRNA seed regions [42].2.13. GeneMANIA: (http://www.genemania.org/). It is a web interface that predicts gene interaction with other genes in various pathways using genetic interactions, pathways, co-expression, co-localization and protein domain similarity [43].
3. Results
- Using dbSNP database for gene data set, the human LH-B gene, gene of our interest, contained a total of 39SNPs missense nsSNPs, 13 nsSNPs of them were predicted to have the most damaging effects on LH-B protein's structure and function.
 | Figure 1. Diagrammatic representation of LH-B gene in silico work flow |
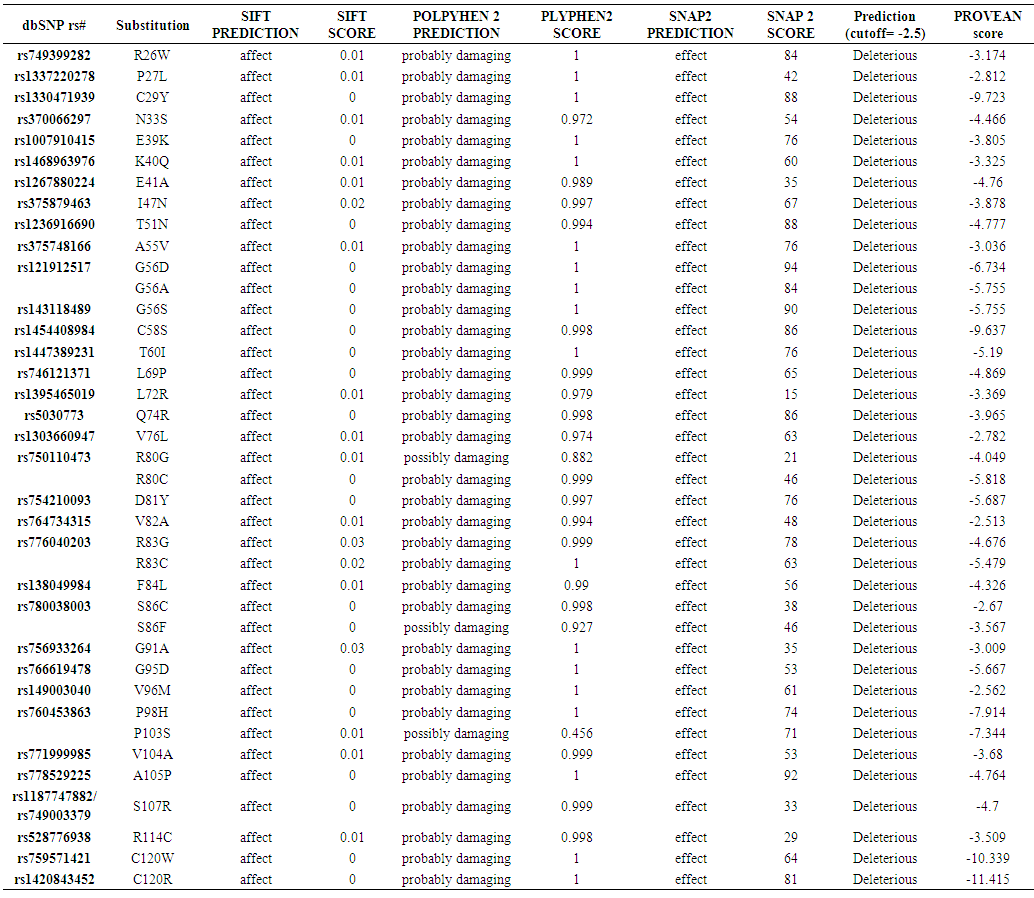 | Table (1). Damaging or Deleterious or effect nsSNPs associated variations predicted by SIFT, Polyphen, PROVEAN, SNAP2 softwares |
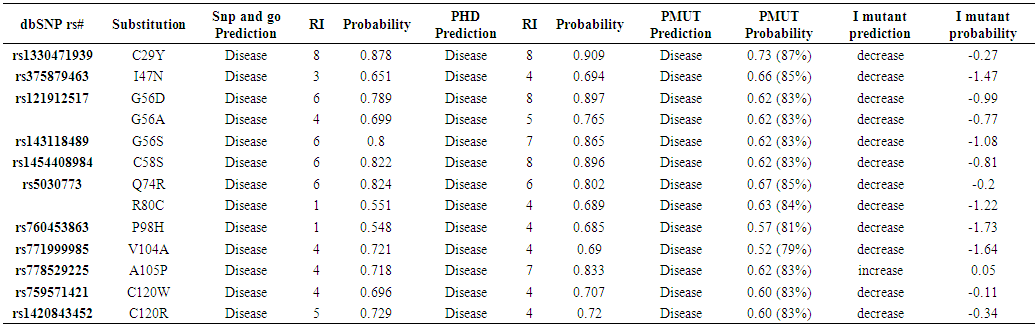 | Table (2). Disease effect nsSNPs associated variations predicted by Pmut, SNP&GO, PHD and I mutant softwares |
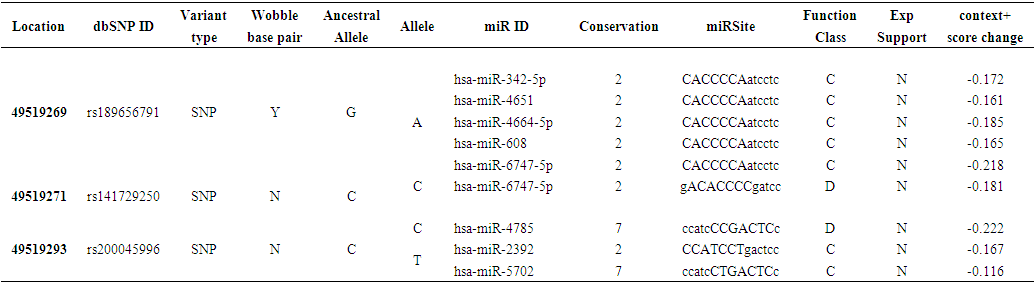 | Table (3). Prediction of SNPs at the 3’UTR Region using PolymiRTS |
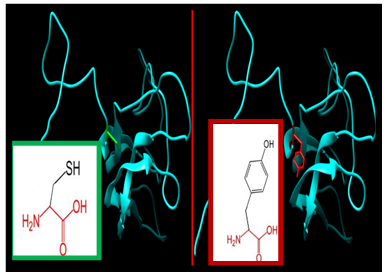 | Figure 2. SNP ID: rs1330471939, (C29Y) The amino acid Cysteine in the native protein (green box) changed to Tyrosine in the mutant protein (red box) at position 29 |
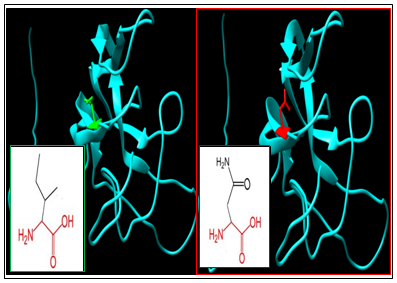 | Figure 3. SNP ID: rs375879463, (I47N) The amino acid Isoleucine in the native protein (green box) changed to Asparagine in the mutant protein (red box) at position 47 |
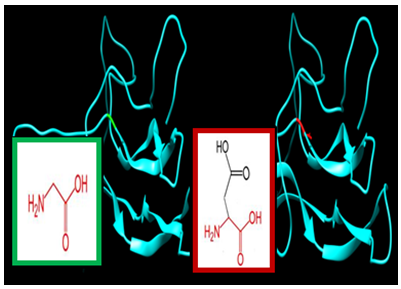 | Figure 4. SNP ID: rs121912517, (G56D) The amino acid Glycine in the native protein (green box) changed to Aspartic Acid in the mutant protein (red box) at position 56 |
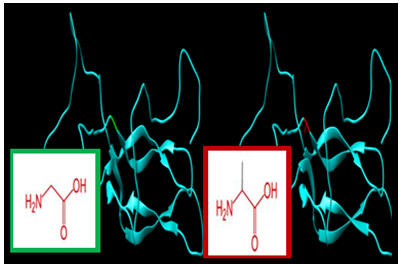 | Figure 5. SNP ID: rs121912517, (G56A) The amino acid Glycine in the native protein (green box) changed to Alanine in the mutant protein (red box) at position 56 |
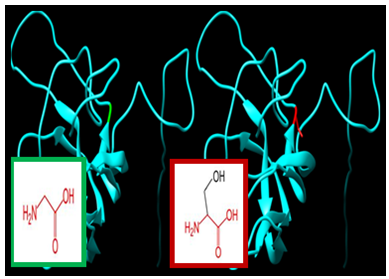 | Figure 6. SNP ID: rs143118489, (G56S) The amino acid Glycine in the native protein (green box) changed to Serine in the mutant protein (red box) at position 56 |
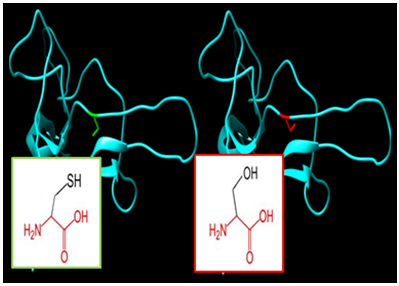 | Figure 7. SNP ID: rs1454408984, (C58S) The amino acid Cysteine in the native protein (green box) changed to Serine in the mutant protein (red box) at position 58 |
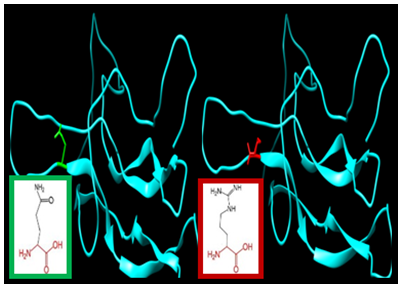 | Figure 8. SNP ID: rs5030773, (Q74R) The amino acid Glutamine in the native protein (green box) changed to Arginine in the mutant protein (red box) at position 74 |
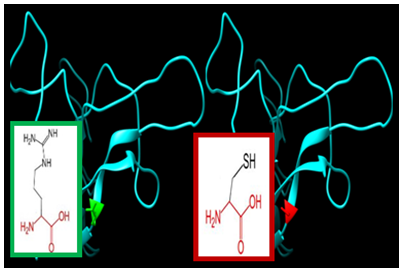 | Figure 9. SNP ID: rs5030773, (R80C) The amino acid Glutamine in the native protein (green box) changed to Arginine in the mutant protein (red box) at position 80 |
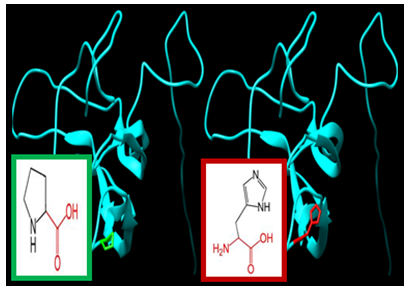 | Figure 10. SNP ID: rs760453863, (P98H) The amino acid Proline in the native protein (green box) changed to Histidine in the mutant protein (red box) at position 98 |
 | Figure 11. SNP ID: rs771999985, (V104A) The amino acid Valine in the native protein (green box) changed to Alanine in the mutant protein (red box) at position 104 |
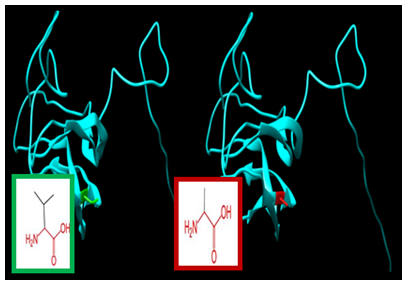
 | Figure 12. SNP ID: rs778529225, (A105P) The amino acid Alanine in the native protein (green box) changed to Proline in the mutant protein (red box) at position 105 |
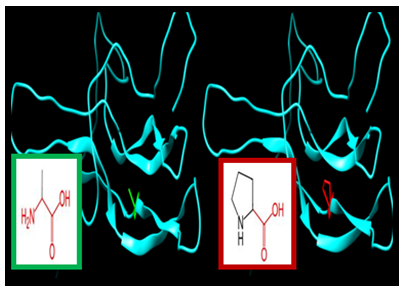
 | Figure 13. SNP ID: rs759571421, (C120W) The amino acid Cysteine in the native protein (green box) changed to Tryptophan in the mutant protein (red box) at position 120 |
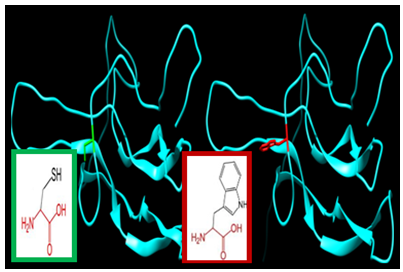
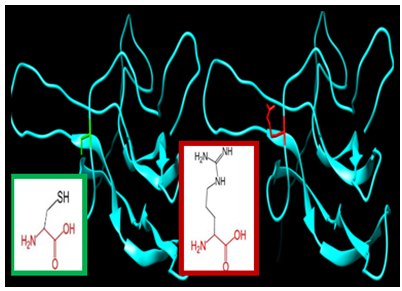 | Figure 14. SNP ID: rs1420843452, (C120R) The amino acid Cysteine in the native protein (green box) changed to Arginine in the mutant protein (red box) at position 120 |
4. Discussion
- LHB gene was investigated in dbSNP National Centre of Biotechnology Information (NCBI public database). This gene contains a total of 404 SNPs in the coding region; of which 140 are missense, 67 synonymous, five nonsense, seven frame shift and 444 are in the non-coding region, of which 300 in the 3'un-translated region (3’ UTR) and 18 in 5’ un-translated region (5’UTR). We selected the missense coding SNPs and 3′UTR SNPs for our investigation.13 novel mutations have been predicated which had an effect on protein stability and function using different bioinformatics algorisms such as SIFT, PolyPhen-2, Provean, SNAP2, SNP&GO, PHD-SNP, P-MUT and I-Mutant 3.0 (Figure 2).Changes in physiochemical properties of the protein due to mutations will possibly result in critical changes in the protein function and consequently result in different phenotypic pictures of the disease.Regarding protein stability, all of the predicted SNPs were predicted to affect protein stability; only one SNP increases protein stability (rs778529225, (A105P)) while the rest decrease its stability. Alteration of one amino acid into another mutated residue results in increasing the size of the protein in different locations such as (rs759571421, (C120W), (rs1420843452, (C120A) and rs760453863, (P98H)) and only two mutations lead to declining the size of it (rs5030773, (R80C) and rs771999985, (V104A)).rs1330471939, (C29Y); In this location, Tyrosine (mutant residue) is bigger than Cysteine the (wild-type residue) which is probably will not fit for cysteine bridge formation which is important for protein stability and this consequently lead to loss of the hydrophobic interactions in the core of the protein. OR This mutation lead to decrease protein stability due to the absence of cysteine bridge, which is important for the stability of the protein and the mutant residue Tyrosine is bigger in size than Cysteine which also results in loss of hydrophobic interactions in the core of the protein.rs375879463 (I47N): alteration of Isoleucine to Asparagine in this location which is nearby to a residue that makes a cysteine bond will affect the binding site of other molecules and loss of hydrophobic interactions in the core of the protein.In position 56; three different mutations occur (rs121912517 (G56D and (G56A) and rs143118489 (G56S)): and affect the physiochemical properties of the protein; all mutated residues lead to decrease stability and increase the size of the protein. Alteration of Glycine to Aspartic Acid with a negative charge which causes repulsion between the mutant residue and neighbouring residues and can disturb interactions with other molecules or other parts of the protein. The torsion angles for this residue are unusual due to the inflexibility of the mutant residue that results in an incorrect conformation and will disturb the local structure of the protein. Moreover, conversion of Glycine to Alanine leads to disturbing interactions with other molecules or other parts of the protein and probably damaging the protein. Also in the same location with other rs143118489 (G56S) Glycine is converted to Serine and results in the formation of unusual torsion angles which probably results into an incorrect conformation and will disturb the local structure of the protein.rs1454408984 (C58S): When Cysteine (wild-type residue) is mutated to Serine (mutant residue) it decreases protein stability due to absences of cysteine bridge and loss of hydrophobic interactions with other molecules on the surface of the protein.rs760453863(P98H): The Histidine (mutant residue) is bigger than Proline the (wild-type residue) but with less hydrophobicity and more flexibility which disturbs the special conformation of the wild-type residue and might cause loss of hydrophobic interactions in the core of the protein.rs771999985(V104A): In this location, the mutation might causes an empty space in the core of the protein because Alanine (mutant residue) is smaller than Valine the (wild-type residue) and cause slightly destabilization of the protein but not totally damage the protein structure. rs778529225(A105P): Only in this location the mutation leads to increase protein stability with bigger size of the mutant residue Proline and probably lead to damage of the protein because it is located near a highly conserved position in the core of the protein.Two mutations occur in position 120 with different consequence on the stability and the neutrality of the protein; one when Cysteine (wild-type residue) is mutated to Tryptophan (mutant residue), rs759571421 (C120W), it decreases protein stability due to absences of cysteine bridge and loss of hydrophobic interactions with other molecules on the surface of the protein. And the other exchange (rs1420843452 (C120A)); when it is mutated to Arginine with a positive charge and bigger size than Cysteine, which causes repulsion between the mutant residue and neighbouring residues because it is located on the surface of the protein and disturbs interactions with other molecules or other parts of the protein.Historically, women with PCOS characterized by a high level of serum LH, high LH/FSH ratio with normal/low FSH level but less is known about the genetic background that leads to overexpression of LH-B gene and its association with PCOS [15-16]. Regarding our findings, two of our most damaging SNPs were reported among the male population suffering from hypogonadism problems associated with low LH level which is uncommon among PCOS population associated with anovulatory and infertility problems [44].Regarding substitution of Glutamine to mutated residue Arginine (rs5030773, Q74R) results in unbinding of luteinizing hormone to its receptors, which consequently, results in failure of puberty to develop spontaneously, with infertility in the homozygous state [45]. In male heterozygotes, the effect of the mutation is more subtle, causing impaired steroidogenesis and a high incidence of infertility, despite the development of normal secondary sex characteristics while a female with the heterogeneous pattern has normal sexual development and fertile which is not agree with the evidence that PCOS associated with high LH level [46-47]. Most reported mutations such as LHβ G1052A and LHβ gene (Trp 8 Arg and Ile 15 Thr) among different populations are with contradictory due to ethnicity and geographical distributions failed to success when analysed by our protocol and did not have an effect on protein structure [19-25]. Therefore, consideration should be taken to these thirteen novel mutations when we are carrying out genetic studies through human samples as the real benefit of computational biology that will contribute in understanding the pathophysiology of the disease that may be useful in discovering new diagnostic and therapeutic markers. Moreover, mutations occur in the 3’UTR region associated with abnormal expression of the gene among target organ explain the abnormal ovulation process which occurs in collaboration with more than 20 genes to accomplish different biological functions. Therefore, damaging its protein due to one of these mutations will probably destroy these pathways.Taking these findings into consideration with the hormonal profiles of women with PCOS will increase our knowledge about the consequences of these mutations that result in overexpression of LH level associated with anovulation problems.
5. Conclusions
- 13 nsSNPs with different positions were predicted to be the most damaging mutations for beta subunit of LH protein, altering its physiochemical properties such as size, charge, hydrophobicity, and stability, leading to loss or disturbance of the protein internal and external interactions and eventually loss of the protein's function as well as disease association. Furthermore, the nine SNPs at the 3UTR were predicted to disrupt miRNA binding sites and hence affect the gene expression function that might be associated with anovulation problems. Uses of in silico analysis tools that allow narrowing the gap between the theoretical and applied parts of the medical research and by computational bioinformatics algorithms, we can increase our knowledge about disease pathogenic process which impacts in di covering newly preventive and treatment strategies tools.
ACKNOWLEDGEMENTS
- The authors desire to acknowledge the excited cooperation of Africa City of Technology-Khartoum, Sudan and to Dr. Mounkaila Noma for language editing and proofreading of the manuscript, University of Medical Sciences and Technology, Khartoum, Sudan.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML