-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Bioinformatics Research
p-ISSN: 2167-6992 e-ISSN: 2167-6976
2017; 7(2): 59-65
doi:10.5923/j.bioinformatics.20170702.02

In Silico Genetic Variation Analysis of Cytochrome P450 2C19 and Their Effect in Certain Drugs Metabolism
Safinaz Ibrahim Khalil 1, Toga A. Alaziz Awad Mahmoud 2, Walaa Salah Abdulla Mohammed 3, Sahar G. Elbager 4, Ahmed M. Elmoselhy 5, Mohamed A. I. Alfaki 6
1Assistant Professor of Pharmacology, Faculty of Medicine, University of Medical Sciences and Technology (UMST), Sudan
2Department of Pharmacology, Faculty of Medical Laboratory Sciences, Sudan University of Science and Technology (SUST), Sudan
3Department of Pharmacology, Faculty of Pharmacy, Sudan International University (SIU), Sudan
4Department of Hematology, Faculty of Medical Laboratory Sciences, University of Medical Sciences and Technology (UMST), Sudan
5Mathematics and Computer Science Department, Faculty of Science Alexandria University, Egypt
6Software Engineering Department, Faculty of Computer Science, Neelain University, Sudan
Correspondence to: Safinaz Ibrahim Khalil , Assistant Professor of Pharmacology, Faculty of Medicine, University of Medical Sciences and Technology (UMST), Sudan.
| Email: |  |
Copyright © 2017 Scientific & Academic Publishing. All Rights Reserved.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Background: Cytochrome P450 2C19 (CYP2C19) is the commonest drug metabolizing enzyme, responsible for the oxidation of about 20% of the currently used drugs. Single Nucleotide Polymorphism (SNPs) play a major role in determining the risk of an individual susceptibility to many illnesses and their drug response. Identification of NPs may help in understanding their effects on genes product and their association with diseases and also could help in the development of new medical testing markers. Methods: This study was carried out for Homo sapiens single variation (SNPs) in CYP2C19 gene through coding regions. Variants data was obtained from dbsnps NCBI database of SNP even. A total of 15943 SNPs in CYP2C19 gene were fond; 14307 SNPs were Homosapins; of which 319 were non-synonymous SNPs. Non synonymous SNPs were selected for insilico analysis; SIFT, Polyphen2, and PharmGKB softwares and servers were used to identify SNPs in proteins functions, structures and, pharmacokinetic activity. Results show that, of 319 nsSNPs, 13SNPs were predicted as highly damaging. Pharmacogenomic analysis of this 13 nsSNPS showed clinical annotations with many diseases and affection of efficacy, pharmacokinetics and toxicity of many drugs. In conclude these previous identified SNPs could lead to gene alteration, which may be contribute to the distinct pharmacogenetic phenotypes.
Keywords: CYP2C19, SNP, dbSNP, Pharm GKB, SIFT, PolyPhen-2, Pharmacogenomic
Cite this paper: Safinaz Ibrahim Khalil , Toga A. Alaziz Awad Mahmoud , Walaa Salah Abdulla Mohammed , Sahar G. Elbager , Ahmed M. Elmoselhy , Mohamed A. I. Alfaki , In Silico Genetic Variation Analysis of Cytochrome P450 2C19 and Their Effect in Certain Drugs Metabolism, American Journal of Bioinformatics Research, Vol. 7 No. 2, 2017, pp. 59-65. doi: 10.5923/j.bioinformatics.20170702.02.
Article Outline
1. Introduction
- The human Cytochrome P450, family 2, subfamily C, polypeptide 19 (CYP2C19) gene located on chromosome ten (10q24), cover approximately 90 kb, is composed of nine exons [1], and its translated enzyme play an critical role in the metabolism of approximately 20% of precribed drugs such as antidepressants, antiulcer, antimalarial, anti-HIV and antiplatelet agents [2-4] About 28 variants alleles in CYP2C19 have been identified [5-8], polymorphism in CYP2C19 and its variants may associated with different pharmacogenetic phenotypes, known as the poor or extensive metabolizers phenotypes results in marked individual variations in the pharmacokinetics and pharmacodynamics and associated with adverse drug reaction or therapeutic failure. [9, 10]Cytochrome P450 2C19 is responsible of the metabolism of many important drugs (Li‐Wan‐Po et al., 2010). Polymorphisms in the CYP enzyme family may lead to the most impact on the metabolism of therapeutic drugs. CYP2D6, 2C19, and 2C9 polymorphisms account almost 80% of drugs in use today are metabolized by these enzymes. CYP2C9 is another clinically significant enzyme that demonstrates multiple genetic variants with a functional implication on the efficacy and adverse effects of drugs (Zhou et al., 2009). In the CYP2C subfamily, the most relevant genetic polymorphism affects CYP2C9 and CYP2C19 genes (Pachkoria et al., 2007). Their adverse drug reactions and therapeutic effects varies among different ethnic groups (Saldana-Cruz et al., 2016). The CYP2C19 enzyme constitute about at least 10 percent of commonly prescribed drugs metabolism and processing, including a drug called clopidogril (1,4). The distribution of common variant CYP2C19 alleles found to vary among different ethnic groups. The allelic frequency of CYP2C19*2 has been shown to be around 17% in African Americans, 30% in Chinese, and around 15% in Caucasians. CYP2C19*3 has been shown to be more among Chinese (5%) and less among African Americans (0.4%) and Caucasians (0.04%). CYP2C19*2 is the dominant defective allele and constitute 75–85% of PM phenotype in Chinese and Caucasian populations (Zhou et al., 2009). Almost all PMs in Asians and Africans can be attributed to CYP2C19*2 and CYP2C19*3. The PM phenotype of CYP2C19 is inherited as an autosomal recessive trait, and the distribution of PM trait varies greatly with ethnic background. (Zhou et al., 2009). The Japanese population incidence of PMs has an 18–23% of S-mephenytoin. Furthermore, 15–17% of the Chinese population are PMs and about 13% of Koreans are PMs. The Asian population has a greater frequency of PM (12–23%), in comparison to Caucasians (1–6%), and black Africans (1–7.5%). The ethnic differences in the metabolism of CYP2C19 substrates are primarily due to the interethnic variation in distribution of the PM trait. The Asian population has around 65–70% PMs and heterozygous EMs, while Caucasians have only 20–25%. The Chinese individuals has a 2-fold greater number of heterozygous EMs than the Caucasian population (Zhou et al., 2009). Many studies have tested the frequency of various CYP2C19 alleles worldwide. The CYP2C19*1 is the wild-type gene is normally referred to as the *1 allele (Li‐Wan‐Po et al., 2010). The existence of these alleles is considered to be predictive for the different phenotypes of the population. Though individuals homozygous for the CYP2C19*2 and CYP2C19*3 alleles are thought to be poor metabolizers (PM), while individuals with at least one CYP2C19*1 allele are justified as extensive metabolisers (EM). Where more recent alleles considered are CYP2C19*17-21, few of which have been functionally identified. Regarding the commonest alleles associated loss of function with increased activity of drugs such as Omeprazole, which become deactivated through this pathway, where prodrugs such as clopidogrel, they are activated through this pathway and loose its activity. The CYP2C19*17 functional effects are unlikely to cause a significant clinical effect only for drugs with narrow therapeutic index (Li‐Wan‐Po et al., 2010). The reported allele frequency of CYP2C9*2 was around 50% in Asians, 18% in Caucasians, 34% in Africans and 19% in American populations. The allele frequency of CYP2C9*3 in the Caucasian, African and Asian populations was <1%, <1% and 7%, respectively. The prevalence of the variant allele was typically <5% in Asians while it is about four times higher in White and African populations (Li‐Wan‐Po et al., 2010). Also, polymorphism within this gene is associated with variable ability to metabolize another drug which is mephenytoin. (2), provided by RefSeq, Jul 2008.Another affected drug metabolism by CYP2C19 therapeutic agents such as the anticonvulsant drug Omeprazole, proguanil, certain barbiturates, diazepam, propranolol, citalopram and imipramine. (3)UniProtKB/Swiss-Prot for CYP2C19 Gene, CP2CJ_HUMAN, P33261.CYP2C19 contribute (>80%) to the metabolism of the Proton pump inhibitors; omeprazole, lansoprazole, and pantoprazole in Extensive metabolizers with a little contribution from CYP3A4 and, probably, CYP2C9. Omeprazole undergoes extensive liver metabolism and the majority of the metabolites formed are 5’-O-desmethylomeprazole, 5- and 3-hydroxyomeprazole, and omeprazole sulfone. Formation of the sulfone metabolite and 3-OH-omeprazole is directed by CYP3A4, while 5-OH-omeprazole and 5’-O-desmethylomeprazole are mostly formed by CYP2C19 (Zhou et al., 2009).Single Nucleotide Polymorphism (SNPs) play a major role in determining the risk of an individual susceptibility to many illnesses and their drug drugs response [11]. Nonsynonymous SNPs (nsSNPs) are one of coding SNPs types, important type of SNPs about 2% of the all known single nucleotide variants; leading to the diversity of encoded human proteins, whereas they affect gene regulation by altering DNA and transcriptional binding factors, maintain the structural integrity of the cell, and affect proteins function [12]. Consequently, identification and analysis of nsSNPs may help in understanding their effects on genes product and their association with diseases and also could help in the development of new medical testing markers and individualized medication treatment [13].Bioinformatics is an important role in almost all aspects of drug discovery, development and assessment. As bioinformatics resources (data bases and software) facilitate the understanding and the prediction of the drug metabolism, especially the following areas absorption, distribution, metabolism and elimination and also toxicity of drugs [2, 14]. Identification of SNPs responsible for adverse drug reaction or therapeutic failure is considered a difficulty, whereas it requires multiple testing for different SNPs in candidate gene. One possible way to overcome this problem was to prioritize SNPs according to their structural and functional significance using different bioinformatics prediction tools. In this study we adopt an insilico approach to analyse human CYP2Cs gene reported mutations using different bioinformatics softwares to investigate the effect of single nucleotide polymorphisms on protein's structure and function and whether these variations can contribute variation in dosage efficacy, toxicity, pharmacokinetic activity or not.
2. Material and Methods
2.1. Datasets
- All the information regarding the SNPs found in human CYP2C19 gene were retrieved using SNPs database (dbSNP NCBI), a genetic variation database established by the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/projects/SNP/). The retrieved data included protein accession number of CYP2C19 gene and all SNPs IDs found in this gene.
2.2. Predicting Damaging Amino Acid Substitutions Using SIFT (Sorting Tolerant from Intolerant)
- Sorting Tolerant from Intolerant (SIFT) is an online bioinformatics software that uses an algorithm to predict the effect of amino acid substitutions, resulting from Non-synonymous SNPs (nsSNPs), on protein function [15]. SIFT uses sequence homology to predict the effects of all possible substitutions at each position in the protein sequence. The score result of each residual ranges from zero to one, where scores close to zero indicate deleterious effect of the substitution, while scores close to one indicate tolerance to substitution [16]. nsSNPs within dbSNP retrieved data were selected as an input for SIFT (http://sift.jcvi.org/).
2.3. Prediction of Functional Modification of Coding nsSNPs Using Polyphen-2 (Polymorphism Phenotyping v2):- PolyPhen-2
- Another software tool that predicts the damaging effects of missense mutations was PolyPhen-2. This tool uses predictive features which involve comparison of a property of the wild-type allele and the corresponding property of the mutant allele. [17] Prediction outcome includes a score that values from 0 to 1 like SIFT with a major difference in which values that are closer to 0 are considered benign while values closer to 1 are assigned as probably damaging. nsSNPs that were predicted to be deleterious by SIFT were submitted to PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/). Then prediction results of the two software’s will then be compared with each other, selecting, i.e. picking out those SNPs who were in common (double-positive results). The results of this output were then rechecked using the SNPs-data base and NCBI-server as well as the existing literature giving us more specific information about the possible responsible gene mutations.
2.4. Evaluation of the of SNPs on the Drug Response. (Pharma GKB)
- The Singapore Pharmacogenomics Portal is the first genomics web platform that provides the opportunity to evaluate the genetic differences among populations for all autosomal genes in the genome, and serves as an integrated platform for linking these data with drugs and genetic variants that affect drug responses, adverse reactions, and dosage requirements. Autosomal genes Annotated as ‘very important pharmacogenes and/or containing variants with high to moderate levels of clinical annotation, level 1A and 2A for CYP2C19 were prioritized (www.pharmgkb.org/downloads, accessed October 2017).
3. Results and Discussion
3.1. Retrieval of SNPs
- The human CYP2C19 gene investigated in this work were retrieved from the NCBI dbSNP database (http://ncbi.nlm.nih.gov/). CYP2C19 gene was containing a total of 15943 SNPs; 14307 SNPs were Homosapins; of which 319 were non-synonymous SNPs. Initially non-synonymous coding SNPs were selected for our investigation.
3.2. Prediction of Protein Structural and Functional Modifications
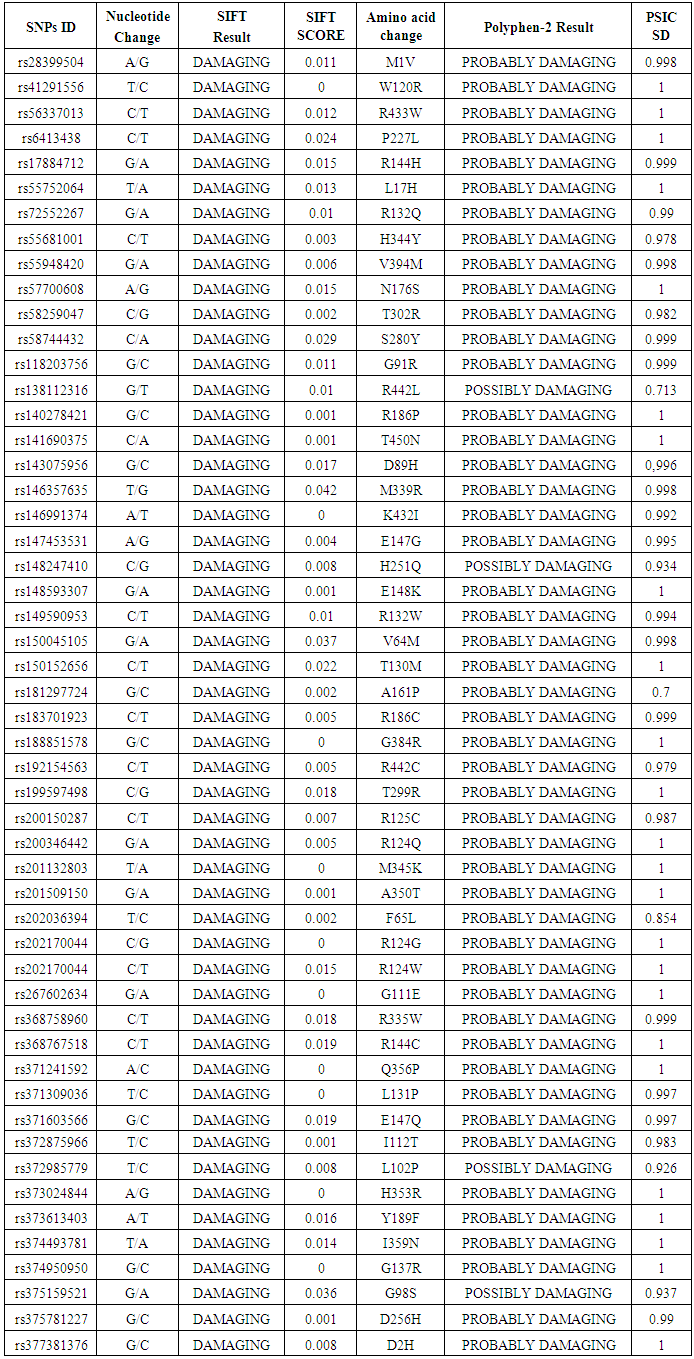
- Coding SNPs were analyzed using SIFT and Polyphen soft-wares. Batch nsSNPs (rs-IDs) were submitted to SIFT server; 69 SNPs were predicted to be deleterious out of 319 SNPs. Deleterious SNPs were submitted to Polyphen-2, 63 SNPs were predicted to be probably damaging, the other 6 SNPs were scored as benign SNPs, 52 variants were predicted to be damaging by both the SIFT and PolyPhen server. 13 SNPs achieved high scores (Tolerance Index (TI) ≤0.005 by SIFT server and PSIC SD=1 by polyphen-2 software) and had been chosen for further analysis (Table 1).
|
3.3. Pharmacokinetics (PharmGKB)
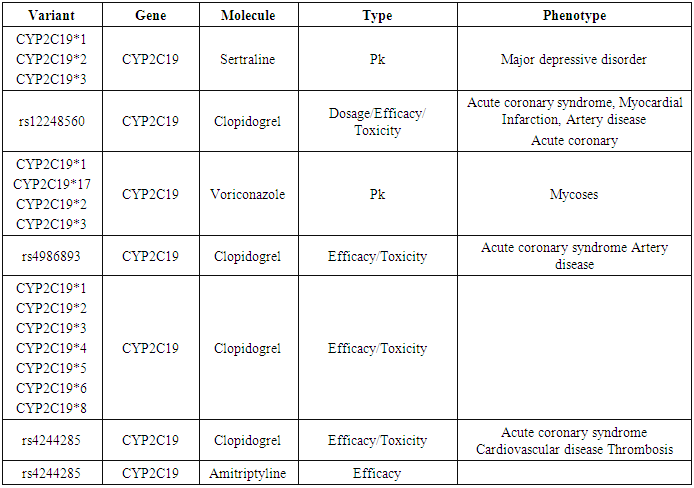
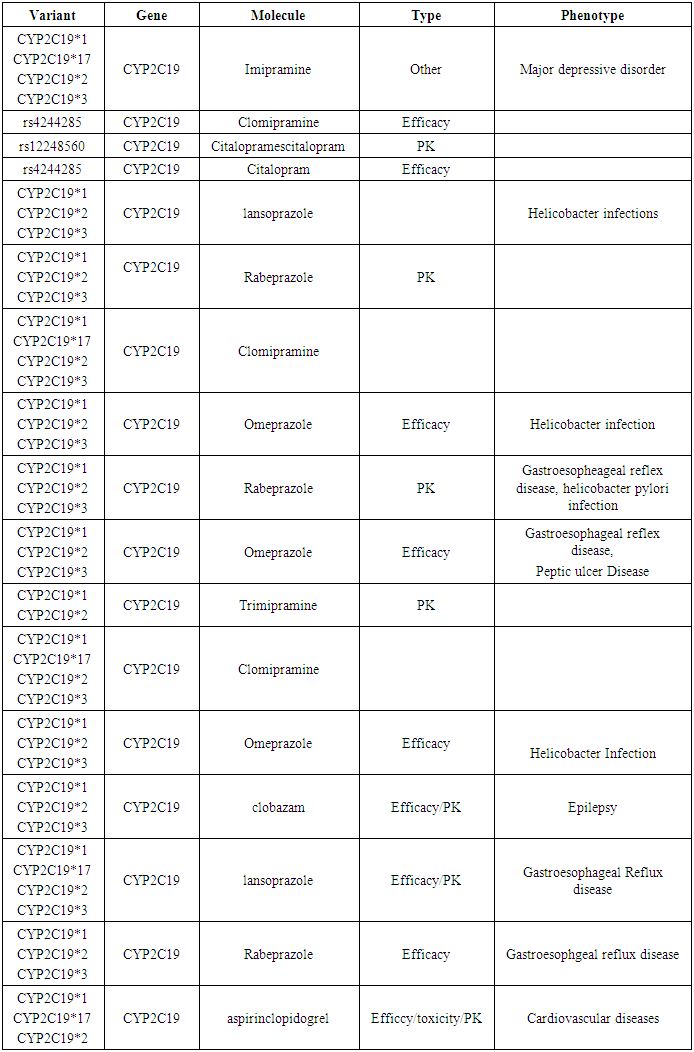
- The SNPS are hypothesized to play an important role in several human diseases. CYP2C19 have many SNPS these result in many interactions with other diseases and drugs affecting their pharmacokinetics, toxicity and efficacy and this predicted by PharmGKB data base. CYP2C19 is annoted as a very important pharmacogene and containing variants with high to moderate levels of clinical annotations PharmGKB (levels 1 And 2) were prioritized.By reviewing the Data base PharmGKB for CYP2C19 gene we found that CYP2C19 *1,2,3,4,5,6,8,19, rs12248560, rs4986893, rs4244285 and rs4244285 were clinically annoted with level 1 and their clinical annotations (Molecule, type of interaction and phenotype): affecting dosage, efficacy, toxicity and pharmacokinetic activity of many drugs such as Sertaline, Clopedogril, Voriconazole and Amitriptyline; affecting many diseases such as Depression, acute coronary syndrome, mycosis and thrombosis (Table 2).
|
|
4. Conclusions
- The CYP2C19 protein is very important causative factor for drug interaction with clinical diseases like Coronary artery disease, Peptic Ulcer Disease, Helicobacter Pylori infection. Our successful Insilco prediction of identification of several pathogenic SNPs in the CYP2C19 gene suggests that the application of computational tools like SIFT, Polyphen- 2, and Pharm GKB may provide an efficiently selecting approach for target SNPs for the conduct of genetic association studies. By analyzing the conformational changes of amino acid residues within CYP2C19 proteins, we have identified 13 nsSNPS to be the most pathological damaging mutations, Pharm GKB analysis of this 13 nsSNPS showed clinical annotations with many diseases and affection of efficacy, pharmacokinetics and toxicity of many drugs. This study demonstrates, the use of insilco tools for application in biomedical research is highly effective and has a great impact on the ability to discover the genetic mutations that cause variation in dosage efficacy, toxicity, pharmacokinetic.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML