-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Bioinformatics Research
p-ISSN: 2167-6992 e-ISSN: 2167-6976
2016; 6(2): 27-31
doi:10.5923/j.bioinformatics.20160602.01

Trans-10,cis-12 Conjugated Linoleic Acid a Novel Inhibitor of Inflammatory Mediators
Rashmi H. K. , Uma Maheswari Devi Palempalli , Sujana Ghanta Vemuri
Department of Applied Microbiology, Sri Padmavati Mahila Visvavidyalayam, Tirupati, India
Correspondence to: Uma Maheswari Devi Palempalli , Department of Applied Microbiology, Sri Padmavati Mahila Visvavidyalayam, Tirupati, India.
| Email: |  |
Copyright © 2016 Scientific & Academic Publishing. All Rights Reserved.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

The present study is designed to understand the interaction between trans-10,cis-12 CLA ligand and the inflammatory mediators like COX-2, iNOS by performing molecular docking studies. Probiotic bacteria such as Lactobacilli are capable of converting linoleic acid to conjugated linoleic acid (CLA). CLA is a collective term used to describe a set of 28 distinct positional and geometric isomers of linoleic acid and the most commonly found isomers are cis-9, trans-11 and trans-10, cis-12 both of which possess biological activity. The trans-10,cis-12 CLA was docked into the COX 2 proteins using CDOCKER CHARMm-based molecular docking algorithm, Accelrys Discovery Studio v 3.5. The binding of COX-2 with CLA isomer revealed that the mode of molecular interaction of trans-10,cis-12 CLA and the standard indomethacin was similar. The in silico results suggest that trans-10,cis-12 CLA isomer could be potent anti-inflammatory agents compared to S-ethyl isothiourea and indomethacin.
Keywords: Anti-inflammatory, Conjugated linoleic acid, Cyclooxygenase-2, Docking, Indomethacin, Inducible nitricoxide synthase
Cite this paper: Rashmi H. K. , Uma Maheswari Devi Palempalli , Sujana Ghanta Vemuri , Trans-10,cis-12 Conjugated Linoleic Acid a Novel Inhibitor of Inflammatory Mediators, American Journal of Bioinformatics Research, Vol. 6 No. 2, 2016, pp. 27-31. doi: 10.5923/j.bioinformatics.20160602.01.
Article Outline
1. Introduction
- Inflammation is an early response to damage instigated by exposure to different pathogens like bacteria, fungi, viruses and a battery of harmful chemical agents which leads to a disrupted tissue homeostasis. Many research reports have demarcated the effect of inflammation in the progression of many diseases such as cancer, atherosclerosis, asthma, and psoriasis. In a majority of these diseases, inflammation has been shown to be a leading cause for pathophysiology of the existing diseases. Chronic inflammation is a long-lasting inflammation that persists for a long time due to continuous stimulus or injury from a persistent pathogen or viral infection. One of the hallmarks of the chronic inflammation is the infiltration of inflammatory cells and fibroblasts leading to prolonged tissue damage [1, 2]. The process of inflammation is mediated by multiple molecular mechanisms; two of the most important are the formation of prostaglandins by cyclooxygenase-2 (prostaglandin Hsynthase) and the production of nitric oxide by the inducible nitric oxide synthase (Figure 1). As per clinical studies and epidemiological studies, ∼ 25% of all cancers are linked to chronic inflammation [3]. Evidence strongly suggests that COX-2 and iNOS play an important role in the development and progression of many tumors [4].
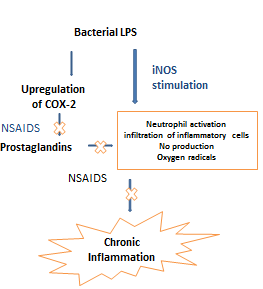 | Figure 1. Inflammatory mediators in chronic Inflammation |
2. Methodology
2.1. Generation of Trans-10,cis-12 CLA Ligand Structure for Docking
- The 3D structure of trans-10,cis-12 CLA (PubChem ID: 5282800) was retrieved from PubChem database. The structure was cross-checked against Zinc database (HTTP://ZINC.dock.org., //ZINC.dock.org., [11]. The resultant structure was optimized and used for docking using CDOCKER of Discovery studio version v 3.5.
2.2. Construction of COX-1 and COX-2 Homology Models
- The amino acid sequence of human COX-2 (P35354.2) with a length of 604 amino acids was obtained from UniProt protein sequence database and it was blasted against Protein Data Bank (PDB) entries to find similar sequences and their structures. The templates for COX-1 and COX-2 were identified as 2OYE (Figure.2A) and 1PXX (Figure.2B) respectively. The COX-1 and COX-2 homology structures were built against the templates structures 2OYE (Ovis aries) and 1PXX (Mus musculus). The 3D crystal structure of 1PXX bound to indomethacin with resolution of 2.90 [Å] was retrieved from PDB. The 3D structure of COX-2 were verified in Accelrys DS with verify protein tool which calculates the likelihood of each residue to be found in its specific local environment and scores the model confirmation using a statistical potential function. The selected model was further analyzed by VERIFY 3D and the energy minimized structures were superimposed with the model structures using Discovery studio visualize [12].
2.3. Docking of CLA Isomers into the Modeled Structures of COX-1 and COX-2
- The trans-10,cis-12 (Figure-3A) was docked into the COX 2 proteins using CDOCKER CHARMm-based molecular docking algorithm, Accelrys Discovery Studio v3.5 (Biosystems Hewlett-Packard, Palo Alto, California, United States). The active site of COX-2 (Figure-3B) protein was defined around the bound inhibitor indomethacin. Inhibitor was removed from the binding site, and chain A was selected for docking. The trans-10,cis-12 CLA isomer was docked against COX-2 and the docking score was compared with standard inhibitor such as indomethacin.
2.4. Docking of CLA Isomer into the Active Site of iNOS
- The protein coordinates of iNOS (PDB_ID:4NOS) was retrieved from Protein data bank (www.rcsb.org). The trans-10,cis-12 CLA and Ethyl-isothiourea (S-Ethyl isothiourea; iNOS-Inhibitor; (PUBCHEM_ID:CID 5139) were docked into the active site of iNOS using CDOCKER [13]. The CHARMm force field was applied along with a grid extension of 8 Å and the partial charges for ligand was set up by Momany-Rone partial charges method. The trans-10,cis-12 CLA isomer along with Ethyl-isothiourea (Figure-3C) was docked with iNOS protein individually into all spheres created around the active sites predicted.
3. Results and Discussion
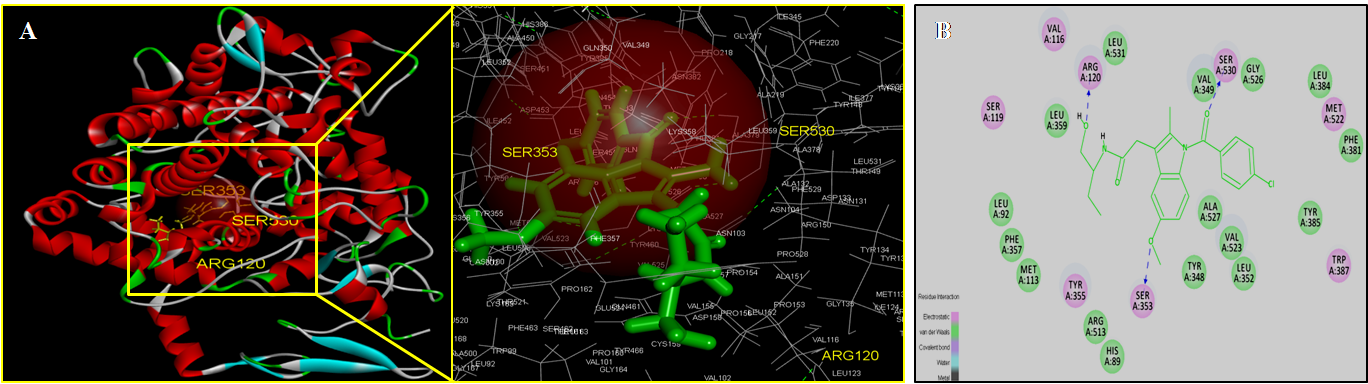
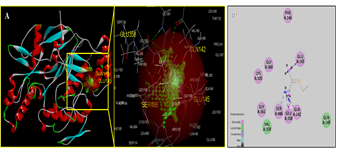
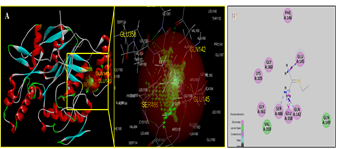
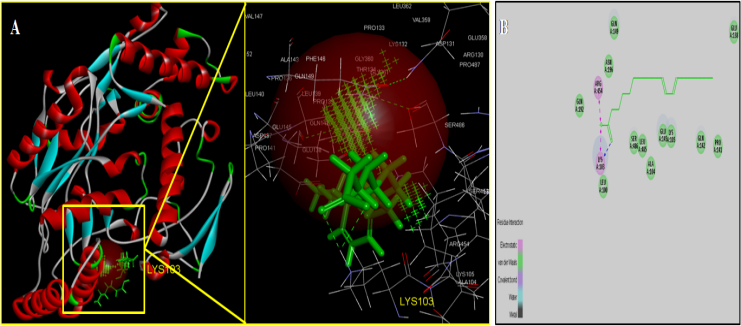
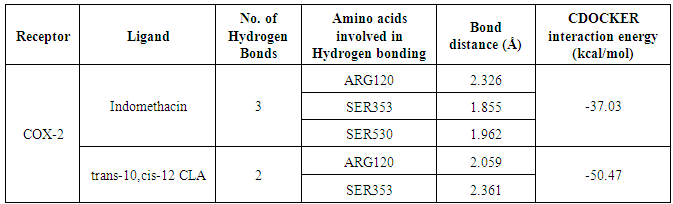
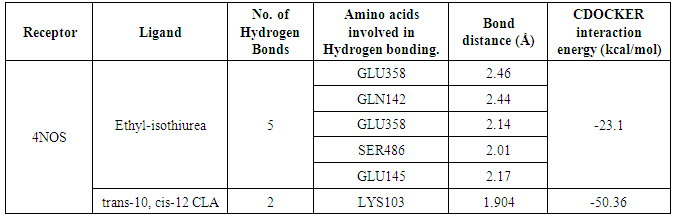
- Molecular docking is a key tool in structural molecular biology and computer-assisted drug design and is useful as it allows a better understanding of molecular events occurring at the binding interface of protein - ligand interaction. It helps in complementing and validating the experimental data. Docking studies of trans-10,cis-12 CLA was carried out to evaluate the most likely target proteins of the inflammatory cascade and to determine the binding mechanism of CLA isomers with target protein using CDOCKER of Accelrys Discovery studio v 3.5.In silico approaches were employed to develop potent COX-2 inhibitors that are attractive and potential leads for various human cancers and inflammatory disorders. The crystal structure of COX-2 could be useful to provide a better understanding about the active sites and the protein-inhibitor binding mechanism [14, 15]. Docking studies were performed using crystal structure of COX-2 complexed with indomethacin. For molecular docking, indomethacin bounded with the crystal structure of COX-2 was removed and docked with trans-10, cis-12 CLA isomer along with indomethacin. The computer docking of standard indomethacin against COX-2 exhibited three H-bonds, one between the carboxyl hydrogen of Indomethacin and ARG-120 with bond distance of 2.32 Å and the second between OH of Indomethacin and SER353 with 1.85 Å of distance and the third bond with OH of Indomethacin and SER350 (Figure-4) whereas trans-10, cis-12 CLA isomer showed two hydrogen bonds with ARG120 and SER353 (Figure-5). The CDOCKER energies with COX-2 for the ligand indomethacin and trans-10,cis-12 CLA were -37.03, - and -50.47, (Table-1). The trans-10, cis-12 CLA isomer used in the present study against COX-2 showed higher binding energy than standard anti-inflammatory drugs indomethacin [15]. As per the earlier reports the carboxylate group of NSAIDS interacts with TYR385 and SER530 and stabilizes the negative charge of the tetrahedral intermediate and demonstrated that TYR385 and SER530 have a structural and functional role in the chelation of NSAIDS ligand. The X-Ray Diffraction structure of human inducible Nitric oxide synthase, PDB_ID 4NOS with 427 amino acid length was considered for docking analysis. Ligands were removed and chain A was considered for docking. Heteroatoms were removed, and polar atoms were added to the structures. The trans-10,cis-12 CLA isomer along with 2-ethyl-isothiourea were docked into the active sites of 4NOS using Discovery studio. Ethyl isothiourea showed 5 hydrogen bonds with GLU358 (2.46Å), GLN142 (2.44 Å), GLU358 (2.14Å), SER486 (2.01Å) and GLU145 (2.17Å) (Figure-6) whereas trans-10,cis-12 CLA isomerexhibited 2 hydrogen bonds with the residue LYS103 (Figure-7). The docking energies of ethyl isothiourea and trans-10,cis-12 CLA isomer with 4NOS were -23.1, and -50.36 kcal/mol respectively (Table 2). However, the binding energies of CLA isomers are higher than ethyl isothiourea.
3.1. Figures and Tables
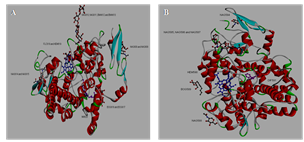 | Figure 2. (A). Homology model structure of COX-1 using the template 2OYE; (B). Homology model structure of COX-2 template with IPXX |
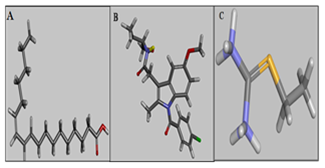 | Figure 3. Structures of Ligands (A) trans10,cis-12 CLA (B) Indomethacin and (C) Ethylisothio urea (Grey-Carbon, Red-Oxygen, Blue-Nitrogen, White-Hydrogen and Yellow-Sulfur) |
|
|
4. Conclusions
- The CLA isomerwas docked into the proteins of inflammatory cascade by using CHARMm-based molecular docking algorithm (CDOCKER), Accelrys Discovery Studio v3.5 (Biosystems Technologies, San Diego, CA, USA) on Z800 workstation. The docking result of COX-2 with CLA isomer revealed that the mode of molecular interaction of trans-10,cis-12 CLA wassimilar to the standard indomethacin. The docking results deduce that trans-10,cis-12 CLA isomer of CLA docked into the same pocket of indomethacin. The CLA isomer demonstrated stronger interaction than standard NSAID indomethacin as docking score is representative of the binding energy of ligand to the receptor. The trans-10,cis-12 CLA isomerwas docked at the predicted active site of 4NOS along with S-ethyl isothiourea, a potent inhibitor of iNOS.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML