-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Bioinformatics Research
p-ISSN: 2167-6992 e-ISSN: 2167-6976
2014; 4(1): 6-10
doi:10.5923/j.bioinformatics.20140401.02
Application of Reverse Vaccinology in Designing a Vaccine for Malaria
Ajao A. T. 1, Adekeye A. O. 2, Yakubu S. E. 2
1Department of Science Lab. Tech., Microbiology Unit, Institute of Applied Sciences, Kwara State Polytechnic, Ilorin, Nigeria
2Department of Microbiology, Ahmadu Bello University, Zaria, Nigeria
Correspondence to: Yakubu S. E. , Department of Microbiology, Ahmadu Bello University, Zaria, Nigeria.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
Malaria is a life-threatening disease and a major public health problem in Nigeria. According to the latest WHO estimates, there about 219 million cases of malaria in 2010 with an uncertainty range of 154 million to 289 million) and an estimated 660,000 deaths mostly among African children. To date, there is no vaccine and the parasites are becoming resistant to available drugs, this necessitates the need for the identification vaccine candidates. Pharmacoproteomic and computational biology represent an attractive alternative approach for the identification of common drug target and peptide-vaccine candidates in the pathogen. Vaccine designing is shifted from entire pathogen or whole antigen to peptide or epitope based-vaccines that are specific, safe and easy to produce. Subtractive genomic approach was used to identify non-human homologous outer membrane proteins in P. falciparum. Four hypothetical proteins were identified based on Vaxijen score and exo-membrane topology, used to produce both B-cell and T-cell mediated immunity. Propred and propred1 were used to predict promiscuos helper T-Lymphocytes (HTL), Cytotoxic T-Lymphocyte (CTL) epitopes and MHCPred for their binding affinity. Three T-cell epitopes derived from identified B-cells bind to maximum number of MHC class I and class II alleles and specifically bind to HLA alleles such as DRB1*0101 and DRB1*0401. The epitopes are LLNNNMLGS, FSVTTNITI and FNVQYAAQL show high potential to induce both B-cell and T-cell mediated immune responses. These predicted epitopes (small peptide) might be promising candidates for vaccine design against malaria. Experimental validation is required.
Keywords: Vaccine, Vaccinology, Epitopes, Malaria, P. falciparum
Cite this paper: Ajao A. T. , Adekeye A. O. , Yakubu S. E. , Application of Reverse Vaccinology in Designing a Vaccine for Malaria, American Journal of Bioinformatics Research, Vol. 4 No. 1, 2014, pp. 6-10. doi: 10.5923/j.bioinformatics.20140401.02.
1. Introduction
- Human race has been struggling against the parasites for centuries and among these parasites. Plasmodium is still one of the most threatening and causing malaria worldwide [1]. It is globally estimated that 243million malaria cases are reported every year. More awe-awakening figures tell us that appropriately 863,000 deaths were caused by malaria in 2008 [2]. Malaria remains one of the most serious health problems worldwide [3] and it is a major public health problem in Nigeria [4]. It accounts for about 60percent of all outpatient attendance and 30percent of all hospital admissions [4]. Malaria increases the morbidity and mortality rates as well as heath problems of developing countries, including Nigeria [5]. In 2001, malaria was ranked the 8th highest contributor to the Global Disability Adjusted Life Year (DALY) and second in Africa [6]. Globally, millions of deaths attributable to malaria are still being recorded. The disease has continued to constitute not only a huge epidemiologic burden in Africa but has also continued to cripple the economic development in the region. The estimated number of fever and malaria episodes per person and year is about 3.5 and 1.5 respectively for children under 5, 1.5 and 0.5 for those 5years and older and a total of 70-110million clinical cases per year. “Malaria causes morbidity through fever, weakness, malnutrition, anaemia, spleen disorder, and vulnerability to other diseases [4]. Despite the fact that malaria is a general phenomenon, children and pregnant women are at greater risk of malaria attack and of suffering long- term after care effects. Evidence from Global Malaria Action Plan [7] suggests that approximately 25millioin pregnant women in Africa are at risk of Plasmodium falciparum malaria and nearly 86 percent of Africa total population is at risk in the endemic areas, therefore, it is a serious problem. For instance in Nigeria, National Malaria Control Programme [8] reported that a child is sick of malaria between 2 and 4 times in a year and it was estimated that malaria was responsible for nearly 110million clinical cases and estimated 300, 000 deaths per year, including up to 11 percentage of maternal mortality. Monetary loss due to malaria in Nigeria is estimated to be about 132million naira in terms of treatment cost, prevention and loss of man-hours [4]. There are four different species of plasmodium: P. falciparum, P. vivax, P.malariae and P. ovale that cause human malaria (18). Among these, P. falciparum is the most mortal [9]. It has high mortality rate as it causes severe complications such as cerebral malaria, renal failure and algid malaria [10]. To date, there are no licensed vaccines against malaria or any other human parasites [11]. The best available treatment, particularly for P. falciparum malaria, is artemisinin-based combination therapy (ACT). If resistance to artemisinnins develops and spreads to other large geographical areas, the public health consequences could be dire, as no alternative antimalarial medicines will be available for at least five years [11]. The search for new targets or vaccine candidates is of high paramount. Bioinformatic-based approach is a novel platform to identify drug targets and vaccines candidates in this parasite. This technique has been successfully used by several authors to identify drug target and vaccine candidates. Peptide – based sub-unit vaccine has recently attracted attention in both treating infectious diseases and also for promoting destruction of cancerous cells [12] these type of vaccines are easy to produce and also safe when compared to the usual vaccines like killed vaccines and attenuated vaccine [12].With the prevailing situation of malaria in this context, this present study was designed to use reverse vaccinology and subtractive genomic approach to identity epitopes that can produce the B-cell and T-cell mediated immunity to develop peptide-based vaccines against malaria.
2. Materials and Methods
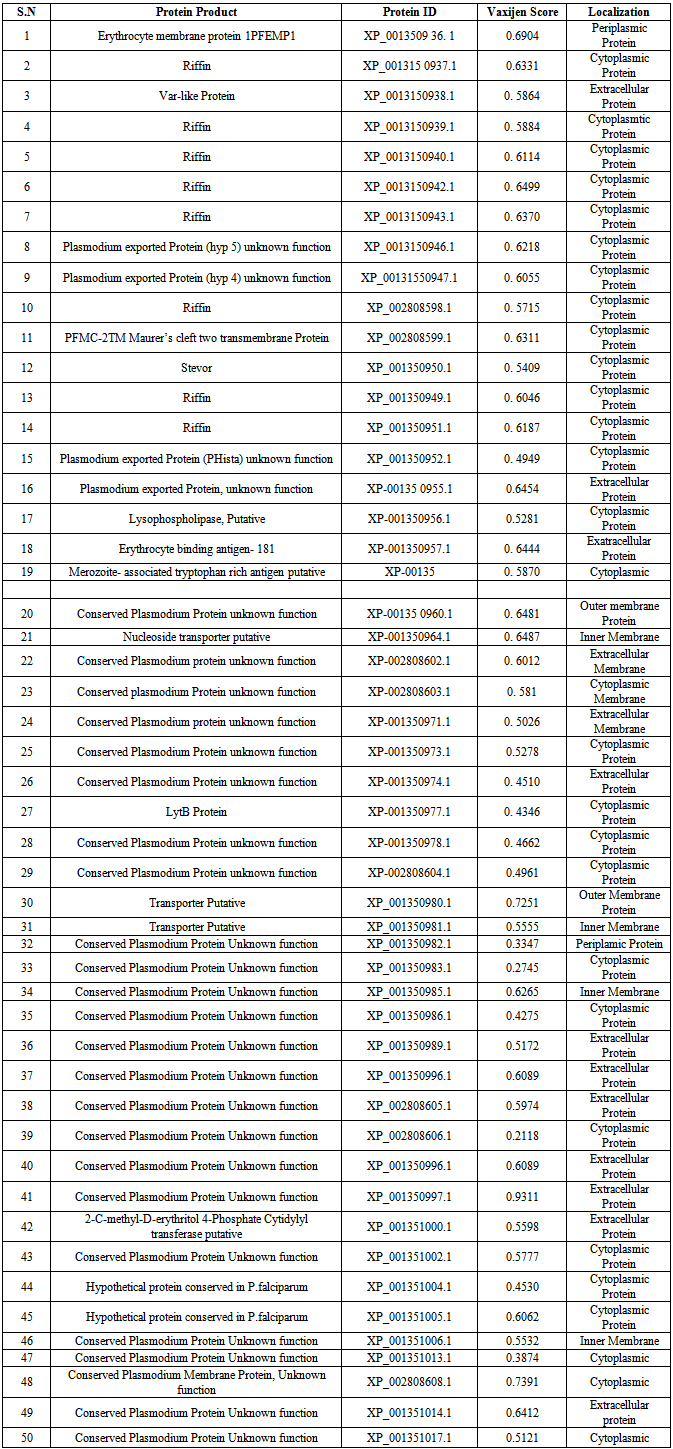
- A complete protein of Plasmodium falciparum was downloaded from National Centre for Biotechnology Information (NCBI) server www.nchi.nlm.nih.gov/genomes. Coding sequences having less than 100 amino acids were not considered. Remaining sequences were subjected to a BLASTP. [13] Search against the non-redundant database with the E-value inclusion threshold set to 10-3. The search was restricted to proteins from Homo sapiens through an option available in the BLAST program. The protein sequences homolog to humans were excluded, while non-human homolog coding sequences (NO Hit found) were listed out as putative therapeutic drug target in Plasmodium falciparum.These non-homologous sequences were subjected to BLASTp analysis against the H. sapiens at an E-value cut off 10-3 [13] BLAST results with No hits with H. sapiens were classified as non-human homologous enzymes. All non-human homologues proteins were then subjected to the program PSORTb V.3 (http.//www.psort.org/psortb/index. Html), for subcellular localization prediction [14]. All the proteins present in the outer membrane were analyzed using vaxijen V 2.0 antigen prediction server(www.Ddg-pharmfac.net/vaxijen). The default parameters (Threshold = 0.4, ACC Output) were used against bacterial species to check the antigenicity of each full length protein sequence. Proteins having antigenic score > 0.6 were selected for B-cell epitope prediction.For prediction of B-cell epitopes each full length protein sequences was subjected to BCPreds analysis (8). Both BCpred and AAP prediction methods [15] of BCPreds were used to identify common B-cell epitopes(http://ailab.cs.iastate.edu/bcpreds). All B-cell epitopes (20mers) having a BCpreds cut off score > 0.7 were selected.Selected B-cell epitopes were then subsequently checked for membrane topology using TMHMM V 2 for exo-membrane amino acid sequences (www.cbs.dtu.dik)Each antigenic B-cell epitope sequences were then analyzed with Propred-1 [16] for MHC Class1 and Propred [16] for MHC Class II epitope prediction using default parameters. Common epitopes for both the MHC classes that also can bind to maximum MHC Alleles were selected and calculated using MHCPred V.2 (Guan et al., 2003) selecting [DRB 1*0101] and vaxijen respectively. Epitopes with highest antigenicity and those bind more than 15MHC molecules comprising of both the MHC class I and II alleles and less than 50nM 1C50 scores for DRB1*0101 were selected.
3. Results and Discussions
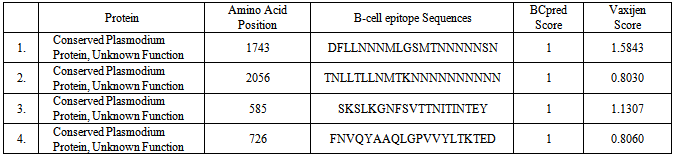
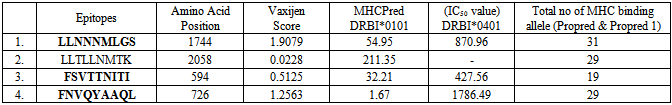
- Falciparum malaria has high mortality rate as it causes severe complications such as cerebral malaria, renal failure and algid malaria [10]. In Nigeria, about half of Nigerian adults have at least one episode of malaria each year and seven out of every 10 patients seen in Nigeria hospitals are ill of malaria [4]. Therefore, Malaria has been put as one of the top killers among communicable disease [17,6]. The present research work used genomic and protein sequences as a basis for pharmacoproteomic studies, driven by the need for malaria control through vaccine and drug development. Earlier studies have reported either T-cell or B-cell based on epitope designing for a given pathogen [18,19,20,21]. An epitope that can produce both B-cell and T-cell mediated immunity is highly desirable for designing peptide based vaccines.A definite homology between the host and pathogen protein chosen as drug targets might lead to unwanted cross-reactions and cytotoxicity [22]. Therefore, enzymes from the P. falciparum that share a similarity with the host proteins were removed to ensure that the targets have nothing in common with the host proteins and thereby eliminating undesired host protein-drug interaction. BLASTP similarity search of all these enzymes at an E-value cutoff of 10-3 resulted in 50 non-homologues enzymes of P. falciparum as shown in Table 1.Neema et al., [22] reported that outer membrane Gram-negative pathogenic bacteria has an important role in the interaction with hosts in bacterial pathogenicity, playing a role in adherence, uptake of nutrients from the host, and countering host defense mechanisms [3]. They could be protective antigens because the components of the outer membrane are easily recognized as foreign substances by immunological defense systems of hosts. The non-human homologous proteins in P. falciparum were analyzed for their localization in outer membrane. The PSORTb server identified non-homologous proteins that reside in the outer membrane. Four membrane associated hypothetical proteins were selected based on their exomembrane topology and vaxijen score to identify epitopes that can induce both B-cell and T-cell mediated immunity are known to be good vaccine candidates [11, 16]. To identify epitopes, full length proteins were subjected to B-cell epitope prediction using BCPreds server. All B-cell epitopes were listed from each protein (Table 2). Epitopes having BCPreds and vaxijen cutoff value respectively > 0.9 and > 0.8 were selected for identification of T-cell epitopes. The common epitopes, that can bind both MHC classes and covers maximum (more than 15) MHC alleles, were selected using propred I and propred servers. In this study, four T-cell epitopes were selected in four selected non-homologous membrane associated proteins. The selected epitopes were further analyzed for vaxijen score and MHCPred IC50 value MHCPred (DRB1* 0101 alleles) was used to identify common T-cell epitopes which can interact with both the MHC classes with highest number and specifically interact with DRB1*01010 as shown in Table 3. The T-epitope must interact with HLA DRB1* 0101 alleles and secure IC50 value not more than 50 which will indicate good binders. [17]. the predicted output is given in units of IC50 nM. A lower value of peptide IC50 indicates higher affinity towards MHC Molecules [12].The four T-cell epitopes: LLNNNMLGS, LLTLLNMTK, FSVTTNITI and FNVQYAAQL were found to be suitable for peptide-vaccine subunit based on total number of MHC score more than 15. Epitope LLTLLNMTK may not be considered as good vaccine candidates in spite, of high value of MHC binding alleles because of its high IC50 value for DRB1* 0101(211.35nM), low vaxijen score with no detectable value for IC50 value DRBI*0401, while the following epitopes can be considered as vaccine candidates: LLNNNMLGS, FSVTTNITI and FNVQYAAQL.ConclusionsThis subtractive genomic and reverse vaccinology approach successfully identified epitopes that can induce both B-cell and T-cell mediated immune responses and these can elicit both humoral and cell mediated immunity. This present finding underscores the utility of large genomic databases for in silico systematic drug target identification in the post genomic era, on the whole, the predicted epitopes, LLNNNMLGS, FSVTTNITI and FNVQYAAQL were antigenic and have much potential to interact with most common human HLA alleles. These might be promising candidates for vaccine design against malaria. However, experimental validation is required.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML