-
Paper Information
- Next Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Bioinformatics Research
p-ISSN: 2167-6992 e-ISSN: 2167-6976
2013; 3(3): 35-41
doi:10.5923/j.bioinformatics.20130303.01
Structural Characterization of the NodD Transcription Factor
Natalia V. Kostiuk1, Maya B. Belyakova2, Dzhulianna V. Leshchenko2, Mikhail V. Miniaev3, Margarita B. Petrova1, Elena A. Kharitonova1
1Biology Department, Tver State Medical Academy, Tver, Russia
2Chemistry and Biochemistry Department, Tver State Medical Academy, Tver, Russia
3Research Center, Tver State Medical Academy, Tver, Russia
Correspondence to: Maya B. Belyakova, Chemistry and Biochemistry Department, Tver State Medical Academy, Tver, Russia.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
Using computer analysis and modeling we studied NodD transcription factor which controls expression of nodulation genes of symbiotic nitrogen-fixing bacteria. On the basis of the amino acid sequences it was predicted that in the secondary structure of NodD protein it must be present ten alpha helices and three beta sheets which form two domains after folding. Since NodD acts in oligomeric mode, it was analyzed all four known for other similar factors of transcription ways of assembly into dimers. Using molecular docking the probable mechanism of NodD activation by binding of low molecular weight inducer (on the model of luteolin) was clarified.
Keywords: Molecular Modeling, Molecular Docking, NodD-protein, Nodulation, Rhizobium
Cite this paper: Natalia V. Kostiuk, Maya B. Belyakova, Dzhulianna V. Leshchenko, Mikhail V. Miniaev, Margarita B. Petrova, Elena A. Kharitonova, Structural Characterization of the NodD Transcription Factor, American Journal of Bioinformatics Research, Vol. 3 No. 3, 2013, pp. 35-41. doi: 10.5923/j.bioinformatics.20130303.01.
Article Outline
1. Introduction
- The formation of nitrogen-fixing nodules is a remarkable example of the symbiotic relationship between the bacteria and leguminous plants. These interactions are highly species-specific: each type of rhizobia complies with a limited host range[1]. At the molecular level, the selectivity is provided by numerous specific determinants. Some of them are specifically secreted by cells, such as plant flavonoids, bacterial lipooligosaccharides (NodD-factor) and proteins. Other specific determinants are integrated into cell membranes[1, 2]. Insight into the molecular mechanisms of symbiotic specificity makes it possible to develop new crop varieties and bacterial strains that could improve the agronomic potential of symbiotic nitrogen fixation[3].Important role in the development of nodules belongs to nod-genes of rhizobia, which are organized in several operons and located either in the chromosome or inSym-plasmids[4]. The expression of many genes is controlled by the NodD transcription factor of nodulation[5]. Its activation occurs in response to the appearance of a plant flavonoid in bacterial cell[3]. Among others, complex flavonoid-protein binds to a promoter region of nodABCIJ operon. As a result of its activation a lipooligosaccharide is synthesized, which is a signal for cell division and the formation of the root nodule tissue[4]. NodD protein is a representative of a numerous family of LysR-type transcriptional regulators (LTTRs)[25].Molecules of these proteins include the two domains: highly conservative DNA binding domain and a variable regulatory domain. LTTRs can form homodimers, homotetramers, and homooctamers[7, 8]. Functionally active tetramer is a result of convergence of two dimers associated with DNA. Usually, the binding sites are located at a distance of 50-60 bp. Joining of LTTR leads to bending of the polypeptide chain which facilitates connection of RNA polymerase and initiation of transcription[7, 9, 10].At present three-dimensional structure of NodD protein remains undeciphered. However, evidence was provided that NodD binds to target DNA through anchoring the two half-sites of the nod box as a tetramer[11, 12]. By experimental and computer methods the complexes of NodD with nodulation genes were studied[13-16]. An imperfect inverted repeat AT-N10-GAT was found in each half-site and is critical for NodD binding[12].DNA binding ability of NodD depends on a number of plant flavonoids such as luteolin, naringenin, eriodictyol, daidzein, and other[3, 17]. However, at the present day the experimental data concerning the localization, structure, and functioning of the binding sites of flavonoids are absent. Also unclear mechanisms of NodD activation under the influence of a low-molecular inductor.The aim of this study was the modeling of spatial assembling of NodD protein and the analysis of structure of binding site for low molecular weight inducer.
2. Materials and Methods
2.1. Analysis of the Amino Acid Sequence Homology
- The amino acid sequence of NodD was found in the database National Center for Biotechnology Information (NCBI). Evaluation of homology as well as identification of other closely related proteins were carried out using software system BLAST[18]. All subsequent studies were performed on the basis of typical sequence of NodD-protein - AAA85282 owned by Rhizobium sp.
2.2. Secondary Structure Prediction
- For the prediction of the secondary structure of the NodD- protein we used several programs based on different algorithms:- PSSFinder[19] – prediction using Markov chains;- PSIPRED[20, 21] - method of neural networks using data obtained from PSI-BLAST- GOR5[22, 23] – Garnier-Osguthorpe-Robson method combines information theory and Bayesian statistics- CDM[24] – combined method based on the method of GOR and the database of protein fragments FDM.
2.3. Modeling of the Tertiary Structure
- Spatial structure was modeled by method of homology (program 3DJIGSAW[25-27], SWISS-MODEL[28-30]) and by ‘threading’ method (the program I-TASSER[31, 32]). In all cases, subunits of 2esn transcription factor were used as template. Additional energy minimization was performed using the Swiss-PDB-Viewer with potential forces of Gromos 96. The residue profiles of three-dimensional models were checked by Verify 3D. The presence of steric and conformational difficulties were assessed using Ramachandran plot and server ProCheck[33]. Residues located in the prohibited regions of Ramachandran plot were not found. Calculation of intramolecular interactions was made by Protein Interactions Calculator[34].
2.4. Docking with a Low Molecular Weight Ligands
- Domain organization of the NodD-protein was studied involving database of Pfam[35]. Search for amino acid residues participated in the formation of flavonoid binding site was implemented by the program I-TASSER and COACH[36-37].For NodD, flavonoids of various structures are low molecular weight ligand[3, 17]. As a model of the ligand we used luteolin. Docking of the ligand to the monomeric and oligomeric proteins was performed by servers SwissDoc[38, 39], PatchDoc[40, 41]. Docked structures with the best estimate were optimized using FineDoc[42, 43].
2.5. Modeling of the Oligomeric Structure
- Obtained in the previous stage model of spatial packing of polypeptides were used to create oligomer. Formation of dimeric structures was performed by servers SymmDock[41, 44], ClusPro[45-47]. Additional energy minimization was carried out using the Swiss-PDB-Viewer. For identifying of steric hindrance, Ramachandran plot and ProCheck were used. The residue profiles were checked by Verify3D.
3. Results and Discussion
3.1. Analysis of the Amino Acid Sequences of NodD
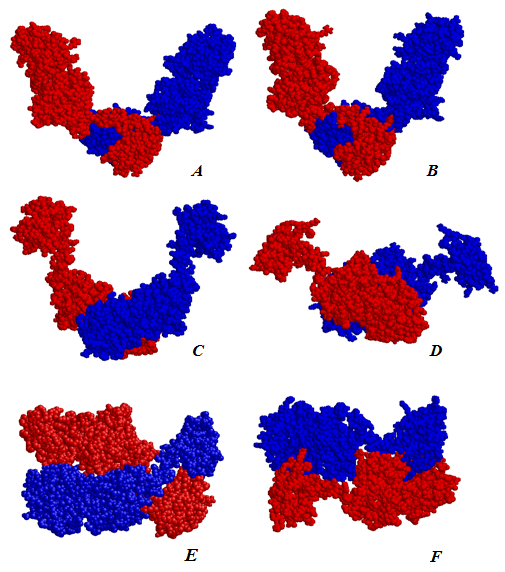
- Currently most sequenced NodD proteins aresingle-stranded molecules of 302-337 amino acids in length. The share of identical amino acids in the orthologs accounts for at least 52%.In order to identify closely related proteins it was performed paired and multiple alignment of studied sequence AAA85282 with other members of the LysR subfamily. 21 proteins were compared: AmpR, CatM, CatR, CynR, CysB, GltC, IciA, IlvY, IrgB, LysR, MetR, NahR, NhaR, NolR, OxyR, PssR, RbcR, SyrM, TcbR, TfdS, TrpI. Multiple alignments did not find a significant similarity within the group. Paired alignments resulted in identification of six proteins with homology with the NodD over 20% (Table 1). In other cases, similar were only short fragments scattered throughout the chain.
|
3.2. The Prediction of Secondary Structure Elements in the NodD Protein
- With a high probability, ten alpha helices extending up to 2.5 turns, and three beta sheets are found in the molecule. The existence of seven other secondary structure elements and their length cannot be determined unambiguously (Fig. 1).
3.3. Modeling of the Structure of Monomeric Protein NodD
- Models of the spatial structure of the monomer NodD obtained by different methods show similar folding motifs but differ in details of structure in irregular regions (Fig. 2). Studied protein is L-shaped with distinct structural domains (Fig. 3). Small N-terminal domain (residues 1-74, the linear size 40 Å × 28 Å × 25 Å) is constructed mainly of spiral elements. In large C-terminal domain (residues 90-308, the linear size 64 Å × 37 Å × 26 Å) the two isolated beta sheets are closed by alpha helices. Domains are connected by a linker region (residues 75-89). In general, the distribution of secondary structure elements matched with predicted by analysis of the amino acid sequence. Calculation of intramolecular interactions showed that the tertiary structure of the protein is supported only by hydrogen, ionic bonds and hydrophobic interactions. Disulfide bonds were not found in the molecule.
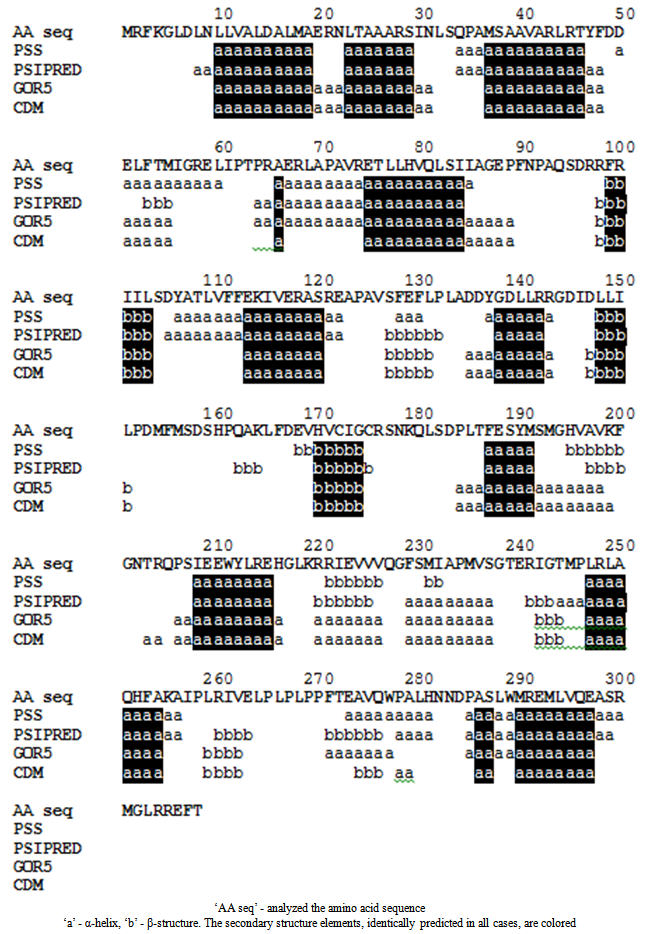 | Figure 1. The prediction of secondary structure elements of NodD |
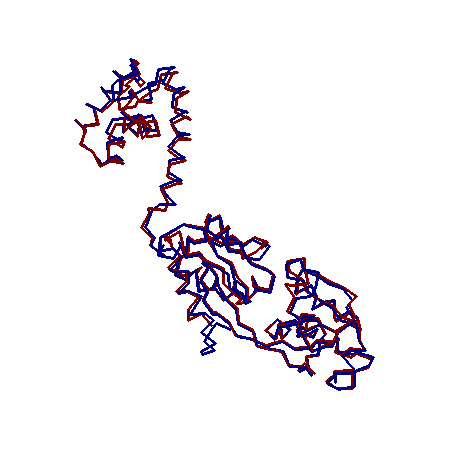 | Figure 2. Superimposition of NodD models: red line - homology modeling, blue line – protein threading |
3.4. Docking of NodD Protein with Inducer
- Additional analysis of protein domain organization involving Pfam database showed a correspondence between structural and functional domains. The N-terminus of the molecule contains hth-domain (‘helix-turn-helix’),responsible for binding with DNA. LysR-substrate domain which provides inductor binding localizes on C-terminal. According to the estimates, the formation of flavonoid binding site may involve amino acid residues 104, 106, 107, 110, 111, 151, 166, 168, 246, 273 (BS-score 1,05). Thus inducer binding site must be located in a recess between two sheets of C-domain (Fig. 3).
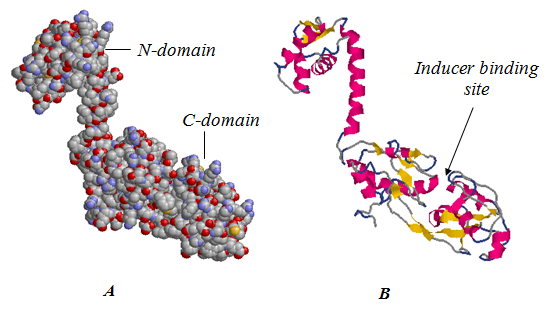 | Figure 3. Probable spatial structure of the NodD protein, presented in different ways: A - ball model B - ribbon model |
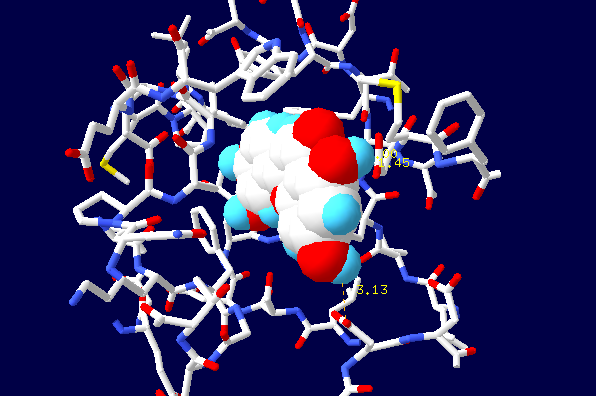 | Figure 4. Location of flavonoid in the binding center |
3.5. Prediction of Oligomeric Structures
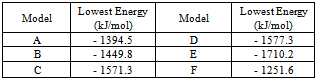
- It is known that bacterial transcription factors LTTF are present in the cell in a variety of oligomeric structures. In a free state homodimers are commonly found. They are responsible for binding with low molecular weight inducer [7]. Functionally active homotetramers arise upon binding to DNA[9,10]. Role of homooligomers is not fully understood [8].Although the domain organization of the LTTF monomers shows a sufficiently high conservatism, formed by them oligomers vary greatly due to structural flexibility. Among dimers there were found V-shaped molecules formed on the basis of contacts between NN and CC domains[48], the molecules with tightly closed subunits connected by principles head-to-head[49] or head-to-tail[50]. Open and closed tetrameric forms were described[51].We analyzed all four variants of dimeric structures known from the literature (Fig. 5, Table 2). The formation of both types of symmetrical V-shaped structures with the N or C domains in the base is energetically allowed (Fig. 5, A-D). In analogy with the transcription factor ArgP[48] we can assume that the first type of dimer interacts with the promoter region of the gene and the second type illustrates possible contact between the C-domain during the functioning of the tetramer. Characteristically, the angle between the subunits may differ noticeably. In the extreme case the complete closure of the subunits is observed (Fig. 5, E). Probably, such flexibility of the structure is required for the functioning of the protein. In the extreme case observed complete closure of the subunits (Fig. 5, E). The binding of low-molecular inductor changes the relative position of the subunits in the dimer. As a result, its attachment to DNA is facilitated. Arising in this case conformational changes make it possible to form a tetramer. The double helix of DNA is bent to facilitate the binding of RNA polymerase. Permissibility of formation of tightly closed head-to-tail dimer draws attention (Fig. 5, F). However, in this case mechanism of its functioning remains unclear.
|
4. Conclusions
- The work performed an attempt to clarify the features of binding of the NodD transcription factor, involved in the regulation of nodulation, with model inducer. Since the spatial structure of the NodD protein still is not solved, we assessed probable elements of protein secondary structure and modeled its tertiary structure. It has shown the possibility of formation of four types of dimers which differ in the type of interaction between the subunits. Probably, such a structural flexibility of NodD is essential to its functioning. Docking of model flavonoid luteolin allowed to determine the position of the binding site of the ligand. Results of the study are important for understanding of the NodD structure. The created model provides a rational basis for the planning of experiments to determine the contribution of different amino acid residues in the formation of the NodD - flavonoid complex. It may be interesting from the point of view of management of NodD activity as well as change of specificity of interaction. The high specificity of such interaction is a serious obstacle in overcoming of species barrier in a symbiotic relationships ‘bacteria-plants’.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML