-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Bioinformatics Research
p-ISSN: 2167-6992 e-ISSN: 2167-6976
2013; 3(2): 11-20
doi:10.5923/j.bioinformatics.20130302.01
Insilico Analysis of Novel hipAB, ccdBA, and yoeB-yefM Toxin-Antitoxin Homolog’s from the Genome of Xenorhabdus nematophila
Jitendra Singh Rathore, Mahendra Pal Singh, Pradeep Gautam
School of Biotechnology, Gautam Buddha University, Greater Noida, Uttar Pradesh, 201308, India
Correspondence to: Jitendra Singh Rathore, School of Biotechnology, Gautam Buddha University, Greater Noida, Uttar Pradesh, 201308, India.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
Xenorhabdus nematophila is a motile gram-negative bacteria belonging to the family Enterobacteriaceae and is a natural symbiont of a soil nematode of family Steinernematidae. The bacterium is essential for effective killing of the insect host and is required by the nematode to complete its life cycle. X. nematophila can be grown under standard laboratory conditions. As the bacterium enters the stationary phase of growth cycle it secretes several extracellular products, which include lipase(s), phospholipase(s), protease(s), and several broad spectrum antibiotics in the insect hemolymph. Recently, the genome of X. nematophila has been completely sequenced and annotated version is available in the NCBI database. In this study the genome of X. nematophila was extensively analyzed bioinformaticaly with NCBI server (www.ncbi.nlm.nih.gov). Our results showed the presence of hipAB, ccdBA, yoeB-yefM toxin-antitoxin homologues at different loci in genome. Later, phylogenetic as well as physiochemical analysis of each toxin-antitoxin pair was done. Extensive promoter analysis of each toxin-antitoxin module was performed with BPROM (www.sofberry.com)to dissect the various transcription factors which may control the transcription of such novel identified putative TA modules in X. nematophila. Existence of all the three operons has been confirmed by polymerase chain reaction (PCR) amplification using operon specific primers.
Keywords: Toxin-antitoxin System, Putative hipAB, ccdBAand yoeb-yefm Operon, Genome, Phylogenetic Analysis, Promoter Analysis, X. Nematophila
Cite this paper: Jitendra Singh Rathore, Mahendra Pal Singh, Pradeep Gautam, Insilico Analysis of Novel hipAB, ccdBA, and yoeB-yefM Toxin-Antitoxin Homolog’s from the Genome of Xenorhabdus nematophila, American Journal of Bioinformatics Research, Vol. 3 No. 2, 2013, pp. 11-20. doi: 10.5923/j.bioinformatics.20130302.01.
Article Outline
1. Introduction
- The term “toxin-antitoxin system” usually abbreviated as “TA system” comprises a functional element consisting of a biologically active protein molecule and a corresponding inhibitor, whose nature and inhibitory mechanism depend on the system’s class affiliation. Components of such systems are encoded within polycistronic operons, often with partially overlapping open reading frames. These systems are wide spread among bacteria as well as archaea[1,2,3,4] and evolved to carry out diverse functions. However, their common feature is an enzymatic activity detrimental for the cell metabolism. Such toxic activity has been demonstrated to switch bacterial cells over to a dormant state, leading to cell death during prolonged exposure. In most cases various stress stimuli are responsible for TA system activation.The signaling pathway in such instances is often related to other stress-induced response pathways. Moreover, it is well documented that in some cases the activity of TA systems stabilizes mobile genetic elements, therefore comprising an important mechanism of plasmids maintenance.The biological activity of a toxin comprising a component of a TA system is usually (but not always) that of an endoribonuclease. The classification of TA systems is based on the mechanism of inhibition of the toxin as well as on operon auto regulatory functions. Initially two classes of TA systems were identified[5], but subsequent discoveries extended the classification to three classes[6]. Class I includes systems in which the antitoxin is an antisense RNA forming duplexes with the toxin mRNA. This leads to inhibition of translation in a process known as RNA interference. Examples of such systems are chromosomally located operons found in Escherichia coli namely tisAB[7] and symER[8], as well as plasmid loci parB[9] of E. coli and par of Enterococcus faecalis[10][11] and a homologous plasmid operon of Staphylococcus aureus[12]. Class II encompasses a wide range of TA systems; Antitoxins of this class are proteins. The biological activities exhibited by the toxins include transcription inhibition by targeting gyrase function and interference with translation through an mRNA interferase activity, which may or may not rely on ribosome binding. The endoribonucleolytic activity of mRNA interferases is often sequence specific gives a short overview of the class II TA systems and their characteristics. Class III comprises a single member only. This system is encoded in the toxIN operon of Erwinia carotonovora, a plant pathogen. In this case inhibition of ToxN toxin activity is driven by RNA molecules directly interacting with the toxin molecules[13][14]. On the basis of different TA Modules, bacteria have developed multiple complex mechanisms ensuring an adequate response to environmental changes. In this context, bacterial cell division and growth are subject to strict control to ensure metabolic balance and cell survival. X. nematophila is a motile gram-negative bacteria belonging to the family Enterobacteriaceae[15]. It forms symbiotic association in the gut of a soil nematode of family Steinernematidae[16]. The X. nematophila isolated, studied so far have been obtained from nematodes harvested from soil samples. Free-living forms of the bacterium have not yet been isolated from soil or water sources, which suggest that the symbiotic association may be essential for the survival of the bacteria in the environment. The bacteria in turn, are essential for effective killing of the insect host and are required by the nematode to complete its life cycle [17][18]. As the bacteria enter the stationary phase of their growth cycle, they secrete several extracellular products, which include lipase (s), phospholipase (s), protease (s), and several broad spectrum antibiotics[19][20]. These products are believed to be secreted in the insects hemolymph when the bacteria enter stationary phase conditions. Since TA modules are involved in the survival of bacterium under stress conditions. Therefore, it could be possible that similar kind of survival mechanism may exist in X. nematophila which protects bacterium during its stress full life cycle inside insect hemolymph or in insect carcass (due to exponential growth of bacterium which leads to nutrient deprivation). In our earlier study we have identified three putative TA modules includes relB, relE and mazF[21] from the genome of X. nematophila. In this study we have identified three additional putative TA modules such as hip-AB, ccd-BA and yoe-yef-BM. These operons located in the genome of X. nematophila, molecular weights as well as isoelectric point of each toxin and antitoxin proteins have been identified with the help of Expasy server (www.expasy.org) which suggests that all three novel identified TA modules in this study belongs to Type II TA category. Extensive promoter analysis were done with BPROM (www.sofberry.com) to dissect the various transcription factors which control the transcription of such novel identified putative TA module in X. nematophila.
2. Methods
2.1. Identification and Genetic Organization of Putative TA Modules
- Complete genome sequence of X. nematophila ATCC 19061 was available in NCBI server (www.ncbi.nlm.nih.gov) under RefSeq database with accession number NC_014228.1. The entire genome sequence of X. nematophila was analyzed bioinformatically for the presence of hip-AB, ccd-BA, and yoeB-yef M homolog’s. Operons coding these TA modules were downloaded and analyzed with ORF (Open Reading Frame) finder Software from NCBI server.
2.2. Protein-Protein Blast
- Protein databank was searched by protein-protein blast performed with putative toxin-antitoxins encoded by the various operon from the genome of X. nematophila using NCBI server (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
2.3. Phylogenetic Analysis
- Different annotated protein sequences were multiple aligned and phylogenetic trees were constructed by using CLC Genomics Workbench (version 4.9) software.
2.4. Physiochemical Properties
- All the physio-chemical properties of putative toxin- antitoxin proteins have been determined by Expasy server (www.expasy.org).
2.5. Promoter Analysis
- Identification of putative pro- moters associated with ORFs was determined by software BPROMO(www.softberry.com).
2.6. Genomic DNA Isolation
- X. nematophila culture was inoculated from glycerol stock in 50 ml and grown overnight at 28℃, 200 rpm. Overnight grown culture was pelleted down by centrifugation at 5000 rpm for 10 minutes at 4℃. The pellet was resuspended in 4 ml TE buffer pH8 (10 mM Tris HCl, 1mM EDTA) and 0.5 ml of 10% SDS was added. 30 μl of proteinase K (20mg/ml) was added to the resuspended culture and incubated at 37℃ for one hour. After complete lysis of the cells, 1 ml of 5 M NaCl was added and mixed gently. 750 μl CTAB NaCl mixture was added to the lysate and incubated at 65℃ for 20 minutes. Later equal volume of chloroform: Isoamyl alcohol mix (approax 7.5ml) was added and mixed gently. It was centrifuged at 12,000 rpm at 4℃ for 30 minutes. To the aqueous phase containing genomic DNA, 12.5 μl of RNase (2mg/ml) was added. The supernatant was incubated at 37℃ for one and half hours; equal volume of phenol: chloroform:isoamyl alcohol mixture was added and mixed properly. The tubes were centrifuged at 12,000 rpm, for 30 minutes at 4℃. The supernatant was again extracted with equal volume of phenol:chloroform:isoamyl alcohol mixture. The supernatant (aqueous phase) was collected in korex tube and 0.6 volume of isopropanol was added and mixed properly. korex tube was centrifuged at 10,000 rpm for 20 minutes at 4℃. The pellet was washed with 70 % ethanol and kept for drying in room temperature. Finally the pellet was dissolved in 0.5 ml autoclaved water and run on 0.8% agarose gel.
2.7. Amplification of Putative HipAB, ccdAB and yoeB-yefM Operon by Polymerase Chain reaction (PCR) from Genome
- The DNA sequence encoding putative hipA toxin and its antitoxin hipB respectively were obtained by PCR amplification using genomic DNA as template (Denaturation at 94ºC for 40 sec, annealing at 60ºC for 40 sec and extension at 68ºC for 1 min 30 sec, total 30 cycles) for cloning with primer pair JSR25 and JSR26. Sequence encoding putative ccdB toxin and its antitoxin ccdA respectively were obtained by PCR amplification using genomic DNA as template (Denaturation at 94ºC for 40 sec, annealing at 60ºC for 40 sec and extension at 68ºC for 50 sec, total 30 cycles) for cloning with primer pair JSR30 and JSR31. Sequence encoding putative yoeB toxin and its antitoxin yefM respectively were obtained by PCR amplification using genomic DNA as template (Denaturation at 94ºC for 40 sec, annealing at 60ºC for 40 sec and extension at 68ºC for 50 sec, total 30 cycles) for cloning with primer pair JSR20 and JSR21.
3. Result and Discussion
3.1. Genetic Organization of hip-AB, ccd-BA, and yoeB-yefM Homolog’s
- X. nematophila genome was analyzed bioinformatically and in this study we have identified three loci corresponding to three different TA modules. HipA toxin homolog encoded by 1299 bp and its antitoxin homolog HipB encoded by 251 bp was located in an operon designated as "XNC1_operon 0810" which lies between 4076692-4077990 base pair of X. nematophila genome in complementary orientation as shown in Figure1 (a).In the second identified putative TA module, putative CcdB toxin encoded by 305 bp whereas its antitoxin homolog CcdA encoded by 278bp. Both genes were located in locus designated as operon"XNC1 _operon0014" which lies between 82145-83430 base pair as shown Figure1 (b). Third identified putative TA module is homolog of yoeB-yefM TA module. Putative YoeB toxin encoded by 254bp whereas its putative antitoxin YefM homolog encoded by 254 bp which is located in operon designated as operon "XNC1_operon0711”. This operon lies between 3607111-3607616 base pair in the genome of X. nemtaophila as shown in Figure 1(c).
3.2. Analysis by ORF Finder (Open Reading Frame Finder) and Identification of Protein Families
- All the corresponding protein sequences encoded by different TA modules were deduced by ORF finder. First putative HipA toxin compose of 432 amino acids and its putative HipB antitoxin composed of 83 amino acids. The Protein-protein blast with HipA protein showed a conserved domain belongs to couple_ hip-A super family, hipA_N super family, and hipA_C super family as shown in Figure 2 (a). Blast results showed its similarity with hip-A domain containing protein of Enterobacter sp.638 with 69% identity (accession no. YP_001176760.1) Similarly, protein-protein blast with HipB protein showed a conserved domain belongs to HTX_XRE super family as shown in Figure 2 (b). The blast results showed its 59% identity with XRE super family transcriptional regulators of Enterobacter sp.638 (accession no. YP_00117676.1).Putative CcdA antitoxin composed of 92 amino acids and its putative toxin CcdB composed of 113 amino acids. Protein-protein blast with putative CcdA protein showed conserved domain belongs to CcdA super family as shown in Figure 3 (a). It showed 68% identity with CcdA protein from Photorhabdus luminescens subsp. laumondii TTO1 (Accession no. NP_ 929535.1). Protein-Protein blast with putative CcdB protein showed a conserved domain belongs to CcdB super family as shown in Figure 3 (b). Blast result with putative CcdB showed that CcdB toxin protein from X. nematophila was 67% identical with CcdB cytotoxic from Photorhabdus luminescens subsp. laumondii TTO1 (Accession no. NP _929536.1)Putative YoeB toxin composed of 84 amino acids and its putative YefM antitoxin composed of 84 amino acids. Protein-protein blast with putative YefM antitoxin protein showed a conserved domain belongs to PhdYefM_antitoxin super family as shown in Figure 4 (a). Blast results showed its similarity with yefM antitoxin protein from Prevent host death protein from Delta proteobacterium MLMS-1. (Accession no. ZP_01288746.1) with 77% identity. Blast results showed that putative YoeB toxin has a conserved domain at N-terminal which belongs to plasmid stability super family as shown in Figure 4 (b). Blast results showed that putative YoeB toxin showed 81% identity with hypothetical protein from Legionella drancourtii LLAP12 LDG_6480 (Accession no. ZP_ 09620091.1). Although putative toxin was encoded by genome but belong to plasmid stability family, this could be possible only due to the horizontal gene transfer commonly present in the prokaryotic system.
3.3. Physiochemical Properties
- The physiochemical properties of putative toxin-antitoxin proteins have been determined by Expasy server (www.expasy.org) which showed that HipA toxin had pI of 6.57 with molecular weight of 48,446 Da whereas its putative antitoxin HipB had pI 10.16 with molecular weight 9480 Da. CcdA antitoxin had pI of 8.89 with molecular weight of 10,546 Da where as its putative antitoxin CcdB had pI 5.09 with molecular weight of 11,543 Da. YoeB toxin had pI of 8.76 with molecular weight of 10,246 Da whereas its putative antitoxin YefM had pI 5.25 with molecular weight of 9512 Da.
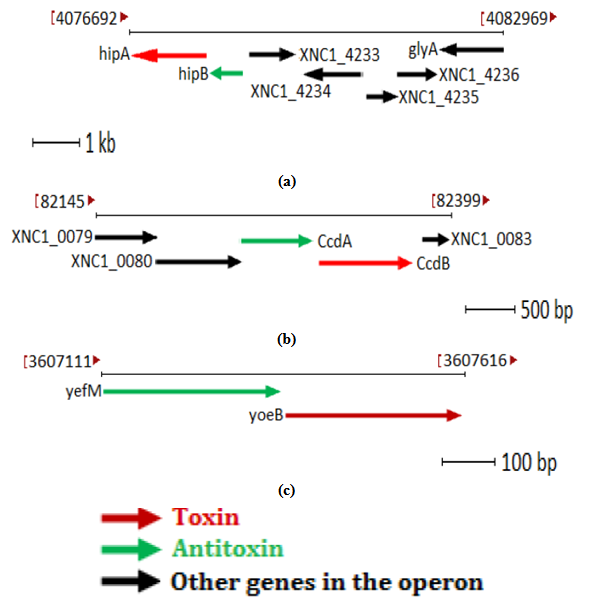 | Figure 1. Genetic organization of TA homolog in the genome of X. nematophila. (a) hipAB TA homolog (b) ccdBA TA homolog (c) yoeB-yefM homolog |
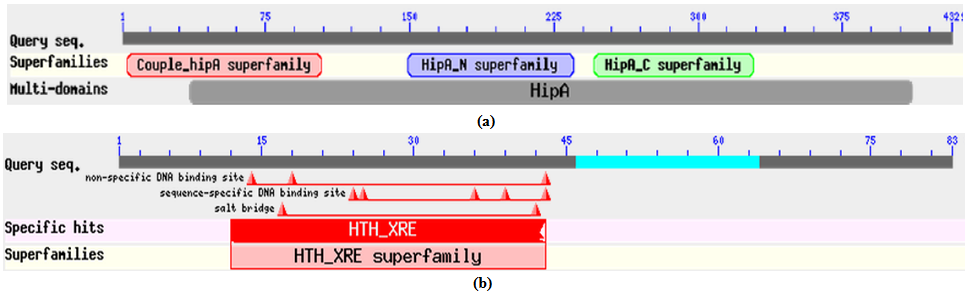 | Figure 2. (a). Conserved domain of putative HipA toxin protein. (b). Conserved domain of putative HipB antitoxin protein |
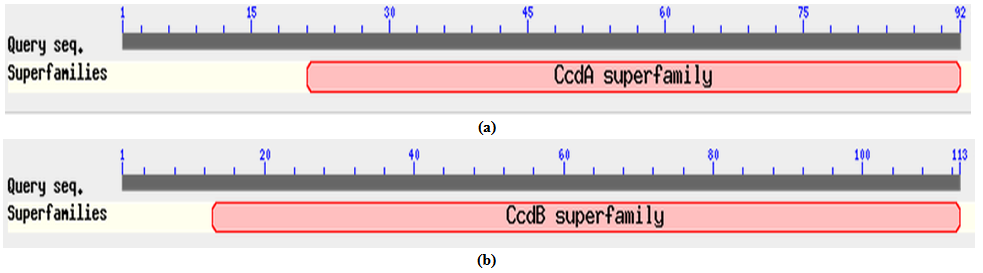 | Figure 3. (a). Conserved domain of putative CcdA antitoxin protein. (b) Conserved domain of putative CcdB toxin protein |
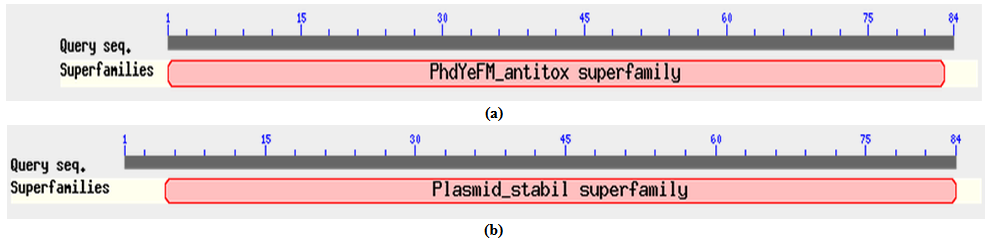 | Figure 4. (a). Conserved domain of YefM antitoxin protein (b) Conserved domain of YoeB toxin protein |
3.4. Phylogenetic Analysis of HipAB, CcdBA, and YoeB-YefM
- Phylogenetic analysis of HipA toxin from X. nemtophila revealed that it forms a distinct branch of toxin from other bacteria as shown in Figure 5. Toxins from other bacteria such as Salmonella enterica subsp. enterica serovar Montevideo str. SARB30 (EHL 47545.1) and Aeromonas hydrophila (YP_0029 95621.1), which are closed to the X. nematophila and located at proximal site in the phylogenetic tree. Whereas toxins from Photobacterium leiognathi subsp mandapamensis svers.1.1. (ZP_08311955.1),Photobacterium angustum S14 (ZP_01233605.1), formed a separate cluster. Rahnella sp. Y9602 (YP_00421 5426.1), Serratia sp. AS12 (YP_0044 98968.1), forms the third cluster located at distal from X. nematophila. Phylogenetic analysis of HipB antitoxin from X. nemtophila revealed that it forms very close related toxin with Enterobacter sp. 638 (YP_001176761.1), and both formed a unique cluster separated from the other two clusters formed by toxins from other bacteria as shown in Figure 6.Phylogenetic analysis of the CcdB toxin revealed that there were three distinct clusters formed by toxins from different bacteria however, toxin from X. nematophila ATCC 19061is very close Photorhabdus luminescens subsp. laumondii TTO1 as shown in Figure 7. Second cluster of toxin has formed from Pectobacterium carotovorum subsp. Brasiliensis PBR1692 (ZP_03827872.1), Citrobacter sp.30_2 (ZP_04558763.1). Third cluster of toxin has formed Klebsiella oxytoca KCTC 1686 (AEX_029 92.1), Escherichia fergusonii B253 (EGC_06526.1). In between second cluster and X. nematophila ATCC 19061, the antitoxin of Yersinia enterocolitica (YP_002643112.1) has separated out evolutionary. CcdA antitoxins from other bacteria such as Yersinia enterocolitica (YP_002643111.1), was separated in very beginning from the putative antitoxin from X. nematophila ATCC 19061 as shown in Figure 8. Similarly another antitoxin from other bacteria such as Citrobacter sp. 30_2 (ZP_04558764.1) separated apart later.
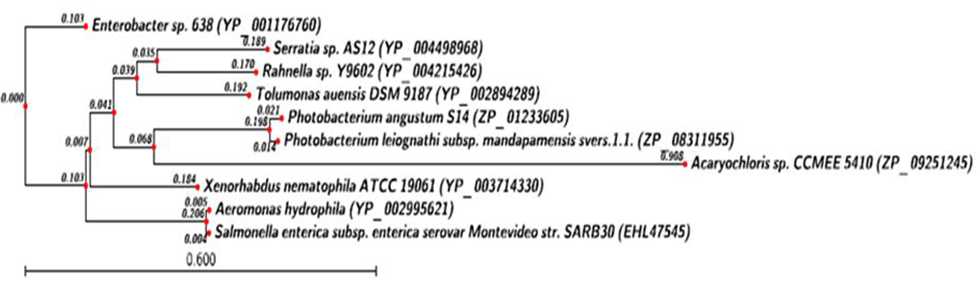 | Figure 5. Phylogenetic analysis of Toxin hip-A protein. Phylogenetic tree constructed based on amino acid sequence of toxin from other bacteria sp. Sequences were aligned by using CLC Genomics workbench (VERSION 4.9) software. Gene accession number for the various protein were as follows: X.nematophila ATCC 19061 (YP_003714330.1), Enterobacter sp. 638 (YP_001176760.1), Salmonella enterica subsp. enterica serovar Montevideo str. SARB30 (EHL47545.1) Aeromonas hydrophila (YP_002995621.1) Tolumonas auensis DSM 9187 (YP_002894289.1) Rahnella sp. Y9602 (YP_004215426.1) Serratia sp.AS12(YP_004498968.1), Photobacterium leiognathisubsp. Mandapamensis svers.1.1. (ZP_08311955.1) Photobacterium angustumS14 (ZP_01233605.1), Acaryochloris sp. CCMEE 5410 (ZP_09251245.1) |
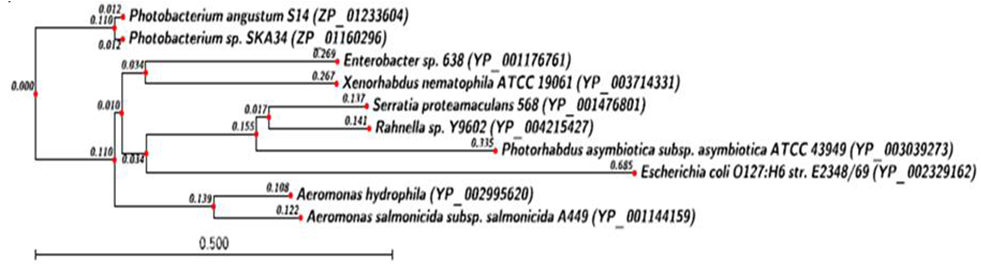 | Figure 6. Phylogenetic analysis of Antitoxin hip-B protein. Phylogenetic tree constructed based on amino acid sequence of antitoxin from other bacterial sp. Sequences were aligned by using CLC Genomics workbench (version 4.9) software. Gene accession number for the various protein were as follows X. nematophila ATCC (YP_003714331.1) , Aeromonas hydrophila (YP_002995620) Aeromonas salmonicida subsp. salmonicida A449 (YP_001144159.1 ), Enterobacter sp. 638 (YP_001176761.1), Photobacterium sp. SKA34 ( ZP_01160296.1), Photobacterium angustums14 (ZP_01233604), Serratia proteamaculans 568 (YP_001476801.1) Rahnella sp.Y9602 (YP_004215427), Photorhadus asymbiotica subsp. asymbiotica ATCC43949 (YP_003039273), Escherichia coli o127:h6 str.e2348/69 (YP_002329162) |
 | Figure 7. Phylogenetic analysis of Toxin ccd-B protein. Phylogenetic tree constructed based on amino acid sequence of toxin from other bacterial sp. Sequence were aligned by using CLC Genomics workbench (version 4.9) software. Gene accession number for the various protein were as follows X. nematophila ATCC 19061 (YP_003710432.1), Photorhabdus luminescens subsp. laumondii TTO1 (NP_929536.1), Salmonella enterica subsp.enterica serovar Paratyphi B str. SPB7 (YP_001591751.1), Yersinia enterocolitica (YP_002643112.1), Dickeya dadantii 3937 (YP_003881572.1), Pectobacterium carotovorum subsp. brasiliensis PBR1692 (ZP_03827872.1), Klebsiella oxytoca KCTC1686(AEX_02992.1), Citrobacter sp. 30_2 (ZP_04558763.1), Providencia rettgeri DSM 1131 (ZP_06124214.1), Escherichia fergusonii B253 (EGC_06526.1) |
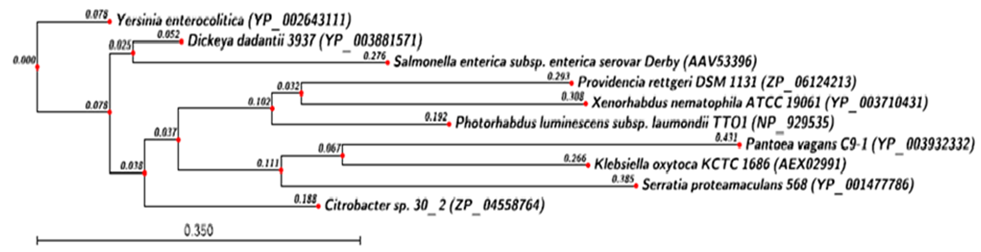 | Figure 8. Phylogenetic analysis of ccd-A Antitoxin protein. Phylogenetic tree constructed based on amino acid sequence of antitoxin from other bacterial sp. Sequence were aligned by using CLC Genomics workbench (version 4.9) software. Gene accession number for the various protein were as follows X. nematophila ATCC 19061(YP_003710431.1), Photorhabdus luminescens subsp. laumondii TTO1(NP_929535), Salmonella enterica subsp. enterica serovar Derby (AAV53396.1), Citrobacter sp.30_2(ZP_04558764.1), Providencia rettgeri DSM 1131(ZP_06124213.1), Dickeya dadantii 3937(YP_003881571.1) Yersinia enterocolitica (YP_002643111.1), Klebsiella oxytoca KCTC 1686 (AEX02991.1),Serratia proteamaculans 568 (YP_001477786.1), Pantoea vagans C9-1(YP_003932332) |
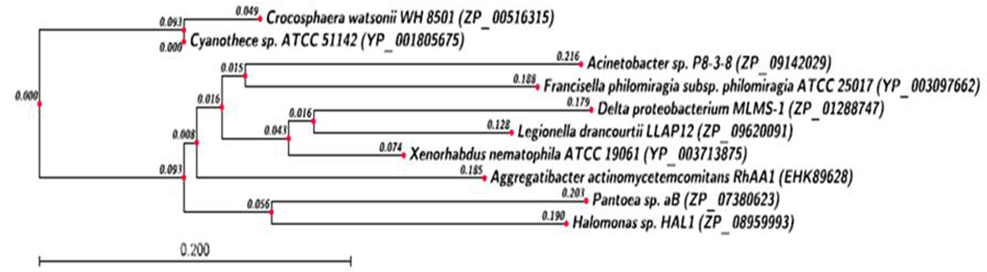 | Figure 9. Phylogenetic analysis of Yoe-B Toxin protein. Phylogenetic tree constructed based on amino acid sequence of toxin from other bacterial sp. Sequence were aligned by using CLC Genomics workbench (version 4.9) software. Gene accession number for the various protein were as follows X. nematophila ATCC 19061 (YP_003713875.1), Legionella drancourtii LLAP12(ZP_09620091) Delta proteobacterium MLMS-1(ZP_01288747.1), Francisella philomiragia subsp. philomiragia ATCC 25017 (YP_003097662.1) Aggregatibacter actinomycetemcomitans RhAA1 (EHK89628.1) ,Cyanothece sp. ATCC 51142 (YP_001805675.1) Acinetobacter sp. P8-3-8 (ZP_09142029.1),Crocosphaera watsonii WH 8501 (ZP_00516315.1), Pantoea sp. aB (ZP_07380623.1),Halomonas sp. HAL1 (ZP_08959993.1) |
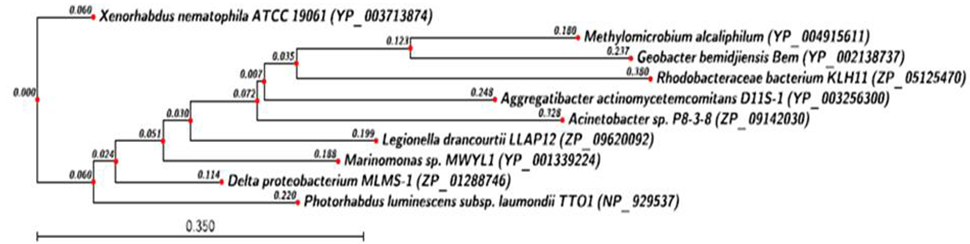 | Figure 10. Phylogenetic analysis of Yef-M antitoxin protein. Phylogenetic tree constructed based on amino acid sequence of antitoxin from other bacterial sp. Sequence were aligned by using CLC Genomics workbench (version 4.9) software. Gene accession number for the various protein were as follows X. nematophila ATCC 19061 (YP_003713874 ), Delta proteobacterium MLMS-1 (ZP_01288746.1) Photorhabdus luminescens subsp. laumondii TTO1 (NP_929537), Legionella drancourtii LLAP12 (ZP_09620092.1), Marinomonas sp. MWYL( YP_001339224.1), Aggregatibacter actinomycetemcomitans D11S-1(YP_003256300.1), Acinetobacter sp. P8-3-8 (ZP_09142030.1), Geobacter bemidjiensis Bem (YP_002138737.1), Methylomicrobium alcaliphilum (YP_004915611.1), Rhodobacteraceae bacterium KLH11 (ZP_05125470.1) |
3.5. Promoter Analysis
- 148 base pairs upstream region to a putative hipAB module contained -35 and -10 promoter like elements along with RpoD17 sigma-D (sigma70) binding sequences as shown in Figure 11. The binding sequence of RpoD17 present in upstream region of putative -35 box and some binding sequences of RpoD17 were overlap with it. RpoD17 is the heat shock sigma factor and is transcribed from two promoters PC and PHS. Synthesis of RpoD mRNA from PC is constitutive under both steady-state and heat-shock growth conditions, while that of PHS is transiently induced upon heat-shock[22]. The putative promoter consensus sequences in the -35/-10 region of the RpoD transcriptional initiation site are highly similar to those of E. coli sigma70. Many bacterial species exhibit a general stress response that can be induced by numerous very different stress conditions and, phenotypically renders the cells broadly stress resistant.
 | Figure 11. 148bp nucleotide sequence upstream region of hip-AB operon showing putative -10 and -35 promoter like elements along with RpoD17 sigma D (sigma 70) binding sequences |
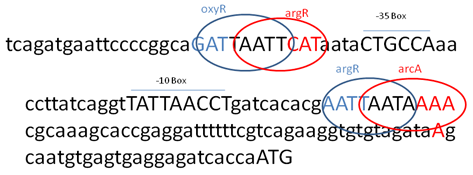 | Figure 12. 152bp nucleotide sequence upstream region of ccd-BA were analyzed. It contained putative -35 and -10 promoter like elements, upstream of -35 box and downstream of -10 box and has oxyR, argR and arcA binding sequences respectively |
 | Figure 13. 143bp nucleotide sequence upstream region of yoeB-yef M were analyzed. It contained putative -35 and -10 promoter like elements, upstream of -35 box and downstream of -10 box and has rpo-D19 and tyr binding sequences respectively |
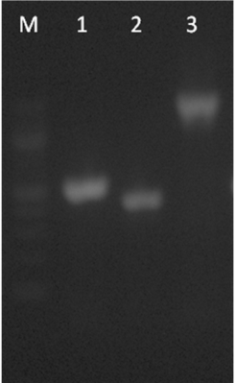 | Figure 14. PCR amplification of ccdBA, yoeB-yefM B and hipAB operon using genomic DNA as template. Lane M, 100bp ladder; lane 1, 585 bp ccdBA operon; lane 2, 505 bp yoeB-yefM operon; and lane 3, 1550 bp hipAB operon |
4. Conclusions
- TA modules have been associated with bacterial programmed cell death and programmed cell survival or persistence[31][32] under various unfavorable conditions which could be physical, chemical or nutrient depleted conditions. X. nematophila encounters competition with soil microflora in insect cadaver as well as with antibacterial protein from the insect hemolymph in its life cycle. Therefore under these conditions, the role of TA modules in X. nematophila cannot be ruled out. In this study, sequenced and annotated genome of X. nematophila has been studied bioinformatically for the identification of putative TA modules in its genome. Three TA module homolog’s hipBA, ccdAB and yoeB-yefM has been identified. Genomic organization revealed that hipAB module is located in “operon="XNC1_operon 0810”. Protein-protein blast of hipA showed its similarity with hip-A toxin from Enterobacter sp.638 and hipB showed its similarity with XRE super family respectively. Therefore, we can presume that in X. nematophila hipA and hipB will form a putative hip-AB TA module. Difference in their individual pI’s (HipA toxin: pI 6.57 and putative HipB antitoxin: pI 10.16) will tend them to form complex in physiological conditions. From the predicted promoter analysis due to the presence of RpoD17 we inferred that transcription of hipA and putative hipB will occur either under different environmental conditions such as high temperature or stress. Similarly, second putative ccdBA TA module is located in “XCN_1operon0014”. This TA module encodes for ccdA antitoxin which showed similarity with ccdA antitoxin protein from Photorhabdus luminesense subs laumondii TT 01, where as its putative CcdB toxin showed similarity with CcdB toxin protein from Photorhabdus luminesense subs. laumondii. In this case also difference in individual pI (Putative CcdB toxin: pI 5.09 and CcdA antitoxin: pI 8.89) will tend them to form complex in physiological conditions. From predicted promoter analysis, presence of oxyR, argR, and arcA indicates that transcription of putative ccdBA module will occur either under SOS conditions and might be tightly regulated. Therefore, we can presume that in X. nematophila gene encoding ccdAB will form a novel TA module. In the third putative yoeB-yefM TA module is located at operon “XCN_1operon0711”. Protein-protein blast with YefM antitoxin protein showed its similarity to the prevent host death protein phd (Antitoxin) from Delta proteobacterium mlms-1 where as YoeB toxin from X. nematophila showed its identity with hypothetical protein from Legionell adrancourtiillap 12 LDG_6480. Therefore, putative YefM antitoxin will form a pair with YoeB toxin. In this case due the differences in individual pI’s (Putative YoeB toxin: pI 8.76 and putative antitoxin YefM pI 5.27) they will form complex in physiological conditions. Promoter predicted revealed the presence of signature sequences for the binding of rpo-D19 and tyr, which indicate transcription form this promoter under heat shock conditions. In silico identification of putative hipBA, ccdBA and yoeB-yefM toxin-antitoxin modules has been confirmed by PCR amplification using genomic DNA as template.
References
| [1] | Mittenhuber G (1999) Occurrence of mazEF-like anti- toxin/toxin systems in bacteria. J Mol Microbial Biotech- nology 1: 295–302 |
| [2] | Gerdes K (2000) Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J Bacterial 182: 561–572. |
| [3] | Pandey DP, Gerdes K (2005) Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res 33: 966–976. |
| [4] | Makarova KS, Wolf YI, Koonin EV (2009) Compre- hensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol Direct 4: 19. |
| [5] | Gerdes K, Wagner EG (2007) RNA antitoxins. Curr Opin Microbial 10: 117–124 |
| [6] | Blower TR, Fineran PC, Johnson MJ, Toth IK, Humphreys DP, Salmond GP (2009) Mutagenesis and functional characterization of the RNA and protein components of the toxIN abortive infection and toxin-antitoxin locus of Erwinia. J Bacterial 191: 6029–6039. |
| [7] | Vogel J, Argaman L, Wagner EG, Altuvia S (2004) the small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr Biol 14: 2271–2276. |
| [8] | Kawano M, Aravind L, Storz G (2007) An antisense RNA control synthesis of an SOS-induced toxin evolved from an antitoxin. MolMicrobiol 64: 738–754. |
| [9] | Gerdes K, Molin S (1986) Partitioning of plasmid R1. Structural and functional analysis of the parA locus. J Mol Biol 190: 269–279. |
| [10] | Greenfield TJ, Ehli E, Kirshenmann T, Franch T, Gerdes K, Weaver KE (2000) The antisense RNA of the par locus of pAD1 regulates the expression of a 33-amino-acid toxic peptide by an unusual mechanism. Mol. Microbial 37: 652–660 |
| [11] | Weaver KE, Reddy SG, Brinkman CL, Patel S, Bayles KW, Endres JL (2009) Identification and characterization of a family of toxin-antitoxin systems related to the Enterococcus faecalis plasmid pAD1 par addiction module. Microbiology 155: 2930–29 |
| [12] | Jensen SO, Apisiridej S, Kwong SM, Yang YH, Skurray RA, Firth N (2010) Analysis of the prototypical Staphylococcus aureus multiresistance plasmid pSK1. Plasmid 64: 135-142 |
| [13] | Blower TR, Fineran PC, Johnson MJ, Toth IK, Humphreys DP, Salmond GP (2009) Mutagenesis and functional characterization of the RNA and protein components of the toxIN abortive infection and toxin-antitoxin locus of Erwinia. J Bacteriol 191: 6029–6039. |
| [14] | Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS, Salmond GP (2009) The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc Natl Acad Sci USA 106: 894–899 |
| [15] | Boemare, N. E., and Akhrust, R. J. 1988. Biochemical and physiological characterization of colony form variants in Xenorhabdus ssp.[Enteriobacteriaceae]. J. Gen. Micro- biol. 134: 751-761 |
| [16] | Herbert, E. E., and Goodrich-Blair, H. 2007. Friends and foes: Two faces of X. nematophila. Nature review. Micro. 5(8): 634-646. |
| [17] | Akhurst, R. J. 1993. Bacterial symbionts of entomo- pathogenic nematodes—the power behind the throne, pp. 127–136. In R. Bedding, R. Akhurst, and H. Kaya (ed.), Nematodes and the biological control of insect pests. CSIRO Publications, Melbourne, Australia. |
| [18] | Kaya, H. K., and Gaugler, R. 1993. Entomopathogenic nematodes. Annu. Rev. Entomol. 38: 181–206. |
| [19] | Akhurst, R. J. 1982. Antibiotic activity of X. spp. bacteria symbiotically associated with insect pathogenic nematodes of the families Hetero- rhabditae and Steinernematidae. J. Gen. Microbiol. 128 (12): 3061-3065 |
| [20] | Nealson, K. H., Schmidt, T. M., and Bleakley, B. 1990. Physiology and biochemistry of Xenorhabdus, p. 271-284. In R. Gaugler and H. Kaya (ed.), Entomo- pathogenic nematodes in biological control. CRC Press, Inc., Boca Raton, Fla. |
| [21] | Jitendra Singh et al.: Insilico Analysis of Novel Relb, Rele and Mazf Toxin-Antitoxin Homolog’s from The Genome of Xenorhabdus Nematophila. American Journal of Bioinformatics Research 2012, 2(3): 21-32 |
| [22] | Aramaki & Fujita, 1999 Recent Insights into the General Stress Response Regulatory Network in Escherichia coli J. Mol. Microbiol. Biotechnol. (2002) 4(3): 341–346.) |
| [23] | Gonzalez F, Gonzalez-Flecha B, Demple B (1997). Transcriptional regulation of the Escherichia coli oxyR gene as a function of cell growth. J Bacteriol 179 (19); 6181-6. PMID: 9324269 |
| [24] | Tao K, Zou C, Fujita N, Ishihama A (1995). "Mapping of the OxyR protein contact site in the C-terminal region of RNA polymerase alpha subunit." J Bacteriol 177(23); 6740 -4. PMID: 7592462 |
| [25] | Caldara06: Caldara M, Charlier D, Cunin R (2006). "The arginine regulon of Escherichia coli: whole-system transcriptome analysis discovers new genes andprovides an integrated view of arginine regulation." Microbiology 152 (Pt 11); 3343-54. PMID 17074904 |
| [26] | Lim87: Lim DB, Oppenheim JD, Eckhardt T, Maas WK (1987). "Nucleotide sequence of the argR gene of Escherichia coli K-12 and isolation of its product, the arginine repressor." Proc Natl Acad Sci U S A 1987; 84 (19); 6697-701. PMID: 3116542 |
| [27] | Charlier92: Charlier D, Roovers M, Van Vliet F, Boyen A, Cunin R, Nakamura Y, Glansdorff N, Pierard A (1992). "Arginine regulon of Escherichia coli K-12. A study of repressor-operator interactions and of in vitro binding affinities versus in vivo repression." J Mol Biol 1992;226(2);367-86. PMID: 1640456 |
| [28] | Kiupakis02: Kiupakis AK, Reitzer L (2002). "ArgR-independent induction and ArgR-dependent superinduction of the astCADBE operon in Escherichia coli." J Bacteriol 184(11);2940-50. PMID: 12003934 |
| [29] | Gunsalus94: Gunsalus RP, Park SJ (1994). "Aerobic-anaerobic gene regulation in Escherichia coli: control by the ArcAB and Fnr regulons." Res Microbiol 145(5-6); 437-50 PMID:7855430 |
| [30] | Engelberg-Kulka, H., and Glaser, G. 1999. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol.53: 43–70. |
| [31] | Amitai, S., Yassin, Y., and Engelberg-Kulka, H. 2004. MazF-mediated cell death in Escherichia coli: a point of no return. J. Bacteriol. 186(24): 8295–8300. |
| [32] | Keren, I., Kaldalu., N., Spoering, A., Wang, Y., and Lewis K. 2004. Persister cells and tolerance to anti- microbials. FEMS Microbiol. Lett. 230:13. |
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML