-
Paper Information
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
American Journal of Bioinformatics Research
p-ISSN: 2167-6992 e-ISSN: 2167-6976
2012; 2(6): 110-118
doi: 10.5923/j.bioinformatics.20120206.02
Study on the Docking Analysis of Homocitrate Synthase Responsible for Hydrogen Production in Nostoc punctiforme ATCC29133
Lakshmi P. T. V. 1, Devi Priyanka 2, A. Annamalai 3
1Centre for Bioinformatics, School of Life Sciences, Pondicherry University, 605014, Puducherry
2Phytomatics Laboratory, Department of Bioinformatics, Bharathiar University, 641046, Coimbatore
3Department of Biotechnology, School of Biotechnology and Health Sciences, Karunya University, 641114, Coimbatore
Correspondence to: Lakshmi P. T. V. , Centre for Bioinformatics, School of Life Sciences, Pondicherry University, 605014, Puducherry.
| Email: |  |
Copyright © 2012 Scientific & Academic Publishing. All Rights Reserved.
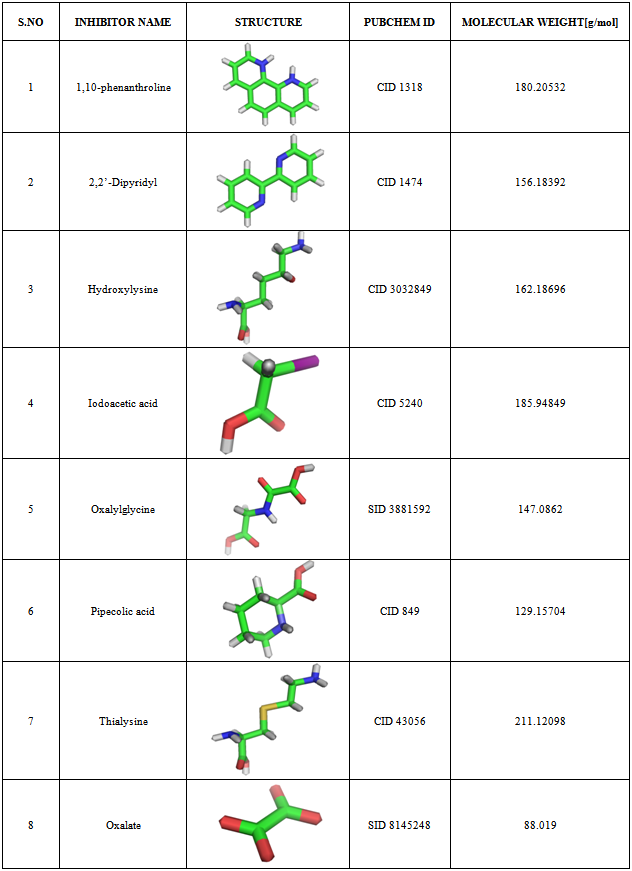
In the case of Nitrogenase-based Hydrogen production, inactivation of the uptake hydrogenase (Hup) leads to significant increase in hydrogen production activity but it lasts only for few hours under the combined nitrogen atmosphere supplied by active hydrogenase. The catalytic FeMo cofactor of nitrogenase binds homocitrate, which is required for efficient nitrogen fixation. Hence blocking of Homocitrate synthase will be an efficient strategy for the significant hydrogen production. Nostoc punctiforme ATCC29133, a nitrogen fixing cyanobacterium that has the genes encoding only for the homocitrate synthase and not for uptake hydrogenase. Since the structure of homocitrate synthase is not yet elucidated, it was taken for modeling using Modeler9v5. The inhibitors for homocitrate synthase were identified from literature and were retrieved from Pubchem. The inhibitors identified were 1,10-phenanthroline, 2,2’-Dipyridyl, Hydroxylysine, Iodoacetic acid, Oxalylglycine, Pipecolic acid, Thialysine, and oxalate were docked with the homocitrate synthase using Argus lab to compare their activities. However, among these eight inhibitors screened, 1,10-phenanthroline and Oxalylglycine were found to have best docking score and proved efficient in terms of both binding affinity and strong hydrogen bond interactions.
Keywords: Nitrogenase, Hydrogenase, Homocitrate Synthase, Nostoc Punctiforme ATCC29133, Modeling, Docking, And Hydrogen Production
Cite this paper: Lakshmi P. T. V. , Devi Priyanka , A. Annamalai , "Study on the Docking Analysis of Homocitrate Synthase Responsible for Hydrogen Production in Nostoc punctiforme ATCC29133", American Journal of Bioinformatics Research, Vol. 2 No. 6, 2012, pp. 110-118. doi: 10.5923/j.bioinformatics.20120206.02.
Article Outline
1. Introduction
1.1. Role of Cyanobacteria in Fuel Production
- Molecular hydrogen, also called the fuel of the future mainly due to its high conversion efficiency, recyclability, and nonpolluting nature, is mostly controlled by either photosynthetic or fermentative organisms and is produced by more environment friendly and less energy-intensive processes as compared to thermo-chemical and electrochemical processes. Cyanobacteria, (blue-green bacteria or blue-green algae) belonging to cyanophyceae, are a large and widespread group of photoautotrophic microorganisms, that fix atmospheric dinitrogen (N2) into ammonia (NH3), a form in which the nitrogen is further available for biological reactions[1]. Although quite uniform in nutritional and metabolic respects, cyanobacteria are a morphologically diverse group with unicellular, filamentous, and colonial forms[2]. It possess several enzymes that are involved in hydrogen metabolism: nitrogenase(s) catalyze the production of hydrogen (H2) concomitantly with the reduction of nitrogen to ammonia; an uptake hydrogenase that catalyzes the consumption of hydrogen produced by the nitrogenase; and a bidirectional hydrogenase, which has the capacity to both uptake and produce hydrogen (Fig.1)[3-4]. In Nitrogenase-based hydrogen production, inactivation of uptake hydrogenase (Hup) leads to significant increase in hydrogen production activity[5]. However, the high-level-activity of the Hup mutants lasts only a few hours under air, a circumstance which seems to be caused by sufficient amount of combined nitrogen supplied by active nitrogenase. While the uptake hydrogenase is present in all nitrogen fixing strains tested so far, the distribution of bidirectional enzyme is not universal (may be present in both nitrogen-fixing and non-nitrogen-fixing cyanobacteria). The molecular masses indicated for the uptake hydrogenase subunits are mean values calculated from the deduced amino acid sequences of Anabaena strain PCC 7120 , Nostoc strain PCC 73102 , and A. variabilis ATCC 29413 , while the values for the subunits of the bidirectional enzyme are based on data exclusively from A. variabilis ATCC 29413 (Tamagnini et al., 2002).
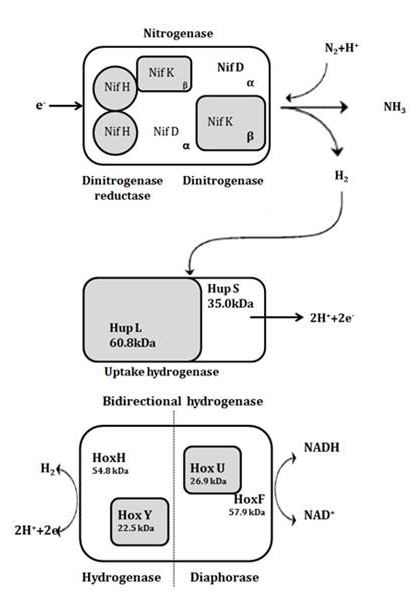 | Figure 1. Enzymes and genes directly involved in hydrogen metabolism in cyanobacteria |
1.2. Biochemistry of the Hydrogen Production
- A metallo-cluster called iron-molybdenum cofactor (Fe-Mo cofactor) is believed to provide the substrate binding and reduction site for biological nitrogen fixation. FeMo cofactor is contained within the nitrogenase MoFe protein, and X-ray crystallographic analysis has revealed that it consists of a metal sulfur core[Fe7S9Mo] and one molecule of (R)-homocitrate. The core is constructed from[MoFe3S3] and[Fe4S3] sub fragments that are connected by three inorganic sulfide bridges located between pairs of Fe atoms from opposing fragments. The catalytic FeMo cofactor of nitrogenase binds homocitrate, which is required for efficient nitrogen fixation[6-10] Homocitrate[(R)-2-hydroxy-1,2,4- butane tricarboxylic acid] is coordinated to the Mo atom through its 2-hydroxy and 2-carboxyl groups[11-13]. Although it is not yet known how the substrate interacts with FeMo cofactor during turnover, the presence of six coordinately unsaturated Fe atoms, as well as the attachment of homocitrate to the Mo atom, has invited speculation about the nature of substrate binding[6]. It has also been proposed that the carboxylate group coordinated to the Mo atom might serve as a leaving group in a mechanism that activates Mo to provide a substrate coordination site[14].
1.3. Role of Homocitrate Synthase
- Homocitrate is synthesized by the nifV-encoded enzyme Homocitrate synthase, which catalyses the condensation of acetyl coenzyme A and 2-oxoglutarate to give homocitrate and CoA. The systematic name of Homocitrate synthase (EC 2.3.3.14) is acetyl-CoA:2-oxoglutarate C-acetyltransferase (thioester-hydrolysing, carboxymethyl forming) while the other names in common use include 2-hydroxybutane-1,2,4- tricarboxylate 2-oxoglutarate-lyase, CoA-acetylating, acetyl-coenzyme A:2-ketoglutarate C-acetyl transferase, and homocitrate synthetase[15]. In fungi, such as yeast, and in extremely thermophilic bacteria, homocitrate is used as an intermediate in lysine biosynthesis through the α- aminoadipic acid pathway[16-17]. This enzyme was found to be a Zn-containing metalloenzyme and belongs to the family of transferases, specifically those acyltransferases that convert acyl groups into alkyl groups on transfer. The R-carboxylate and R-oxo groups of R-ketoglutarate are perhaps required for optimum binding to coordinate to the active site Zn[18]. However, its activity can be inhibited by some of the metal-chelating and sulfhydryl binding agents such as 1,10-phenanthroline, Oxalylglycine, 2,2’-Dipyridyl, Oxalate and Iodoacetic acid[19-20]. Moreover, in the in vivo condition, the activity of Homocitrate synthase was inhibited by Lysine and its analogues such as Hydroxylysine and Thialysine, which had a heterotrophic effect on 2-ketoglutaric binding sites. Perhaps, a second class of affectors found, included 2-aminoadipic acid, pipecolic acid and dipicolinic acid, which affected the S-acetyl coenzyme A binding sites in Saccharomyces lipolytica[21].
1.4. Criterion for Choosing the Organism
- Nostoc punctiforme ATCC29133 (Nostocaceae) is a nitrogen-fixing cyanobacterium that grows autotrophically with CO2 as the carbon source, utilizing an oxygen-producing photosynthetic mechanism for the generation of ATP and reductant. Interesting, in this organism, heterocyst occupies 3-10% of total cells, which differentiate in response to the lack of nitrogen in the environment and acts as the site of nitrogen fixation. A latest release of Cyanobase (http://genome.kazusa.or.jp/cyanobase/NPUN), a genome database for Cyanobacteria, has revealed Nostoc punctiforme ATCC29133 to have the genes encoding solitarily for Homocitrate synthase and not for Uptake hydrogenase. And moreover, the Kyoto Encyclopedia of Genes and Genomes metabolic pathway database also suggested that lysine biosynthesis in Nostoc sp. to proceeds by the diaminopimelic acid pathway, which does not involve homocitrate instead of α-aminoadipic acid pathway, which does involve homocitrate[5]. Hence blocking of Homocitrate synthase would be an efficient strategy for the significant hydrogen production[17].
1.5. Objective
- With this perspective, the present study was aimed to work on cyanobacterial enzyme alteration to enhance the production of Hydrogen. Since, comparative modeling is a useful tool in bioinformatics to predict the three dimensional structure of an unknown protein and also aids to understand protein function, the present study was aimed in this direction to model and predict the structure of Homocitrate synthase using Modeller 9v5 for the sequence (Accession No: B2J701) of Nostoc punctiforme ATCC29133, which was obtained from UniProt database. Moreover, the interaction between Homocitrate synthase and its potential inhibitors such as 1,10-phenanthroline, 2,2’-Dipyridyl, Hydroxylysine, Iodoacetic acid, Oxalylglycine, Pipecolic acid, Thialysine, and Oxalate was also investigated Insilico by the process of docking.
2. Methodology
2.1. Retrieval of Target Protein Sequence
- The protein sequence of homocitrate synthase from Nostoc punctiforme ATCC29133 was obtained from the Uniprot database (Accession No: B2J701) (http://www. uniprot.org/uniprot/B2J701). Since the three dimensional structure could not be ascertained in PDB (http://www.rcsb. org/), an attempt was made to determine the 3D structure.
2.2. Template Identification
- The NCBI BLAST was used to identify the template for modeling the three dimensional structure of Homocitrate synthase of Nostoc punctiforme ATCC29133, which revealed no suitable template identity. Therefore multiple templates (3BLE_A, 2CW6_A, 3BG9_B) were identified using 3D-JIGSAW(http://bmm.cancerresearchuk.org/~3djigsaw/).
2.3. Model Generation and Validation
- The three dimensional structure of Homocitrate synthase was predicted using MODELLER9V5 (http://www.salilab. org/modeller/) where the script "align2d.py" was employed to perform an alignment between the target and template sequence and the script "model-default.py" was employed to obtain a rough 3D model which was subjected to energy minimization using the steepest descent technique and Conjugate gradient technique to eliminate bad contacts between protein atoms through Swiss-Pdb Viewer (http://expasy.org/spdbv/). Perhaps, the backbone conformation of the rough model was inspected using the Phi/Psi Ramachandran plot obtained in the PROCHECK server (http://nihserver.mbi.ucla.edu/saves/).
2.4. Active Site Prediction
- After obtaining the final model, the possible binding sites of Homocitrate synthase was searched using Q-site Finder (http://www.modelling.leeds.ac.uk/qsitefinder/) which works by binding the hydrophobic (CH3) probes to the protein, and finding clusters of probes with the most favorable binding energy. The higher cavity cluster was considered and the residues around the cluster were identified as the binding residues using Pymol. Thus, ten binding sites were obtained for Homocitrate synthase and since, the amino acid present in the first active site formed the binding pocket forming a hollow sphere arranged accordingly to the binding of Zinc metal was chosen to study interactions at this site with the chosen ligands.
2.5. Protein- Ligand Interaction Studies
- Based on the previous studies[18-21] in microorganisms such as Saccharomyces cerevisiae and S. lipolytica, the following eight compounds viz., 1,10-phenanthroline; 2,2’-Dipyridyl; Hydroxylysine; Iodoacetic acid; Oxalylglycine; Pipecolic acid; Thialysine; and Oxalate were found to effectively inhibit Homocitrate synthase, of which 1,10-phenanthroline; 2,2’-Dipyridyl; Oxalylglycine and Oxalate were metal chelating agents; while Iodoacetic acid belongs to the sulfhydryl- binding agent; Hydroxylysine and Thialysine belongs to lysine analogues; and Pipecolic acid -an affector were chosen as the inhibitory ligands. The 3D structure of the inhibitors along with the Pubchem ID and Molecular weights (http://www.ncbi.nlm.nih.gov/Structure/ index.shtml) were determined (Table 1) in order to dock with homocitrate synthase in Argus lab 4.0 (http://www.arguslab. com/downloads.htm), with the grid resolution of 0.40 Å where flexible ligand docking was done when the ligand was described as a torsion tree and the grids were constructed to overlay the binding site. Ligands root node (group of bonded atoms that do not have rotatable bonds) was placed on a search point in the binding site and a set of diverse and energetically favourable rotations were created. For each rotation, torsions in breadth-first order were constructed and those poses that survive the torsion search was scored. The N-lowest energy poses were retained and the final set of poses that underwent coarse minimization, re-clustering, and ranking was selected. The file of the receptor and ligand were uploaded in pdb format and mol format respectively. Docking precision was set to ‘Regular precision’ and ‘flexible ligand docking’ mode was employed for each docking run. The best ligand-receptor structure from the docked structure was chosen based on binding affinity and strong hydrogen bond interactions.
3. Results
3.1. Homology Modeling of Homocitrate Synthase
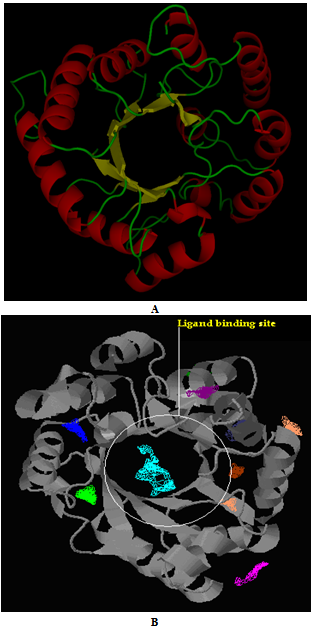
- Homocitrate synthase of Nostoc punctiforme ATCC29133 constituted 377 aminoacids with the molecular weight of 41 kDa. The absence of three dimensional structures in PDB prompted to construct the 3D model, which provided valuable insight into molecular function and also enabled the analyses of its interactions with suitable inhibitors. Among the four conformations generated, the one with the least modeller objective function value was considered to be thermodynamically stable and chosen for further refinement and validation (Fig 2A). The stereochemistry of the constructed model was subjected to energy minimization to assess the quality of the structure. Ramachandran plot for the model showed 84.9% of the residues in the core region, 14.4% residues in the allowed regions and 0.6%, that is two residues in the disallowed region (Fig 3).
|
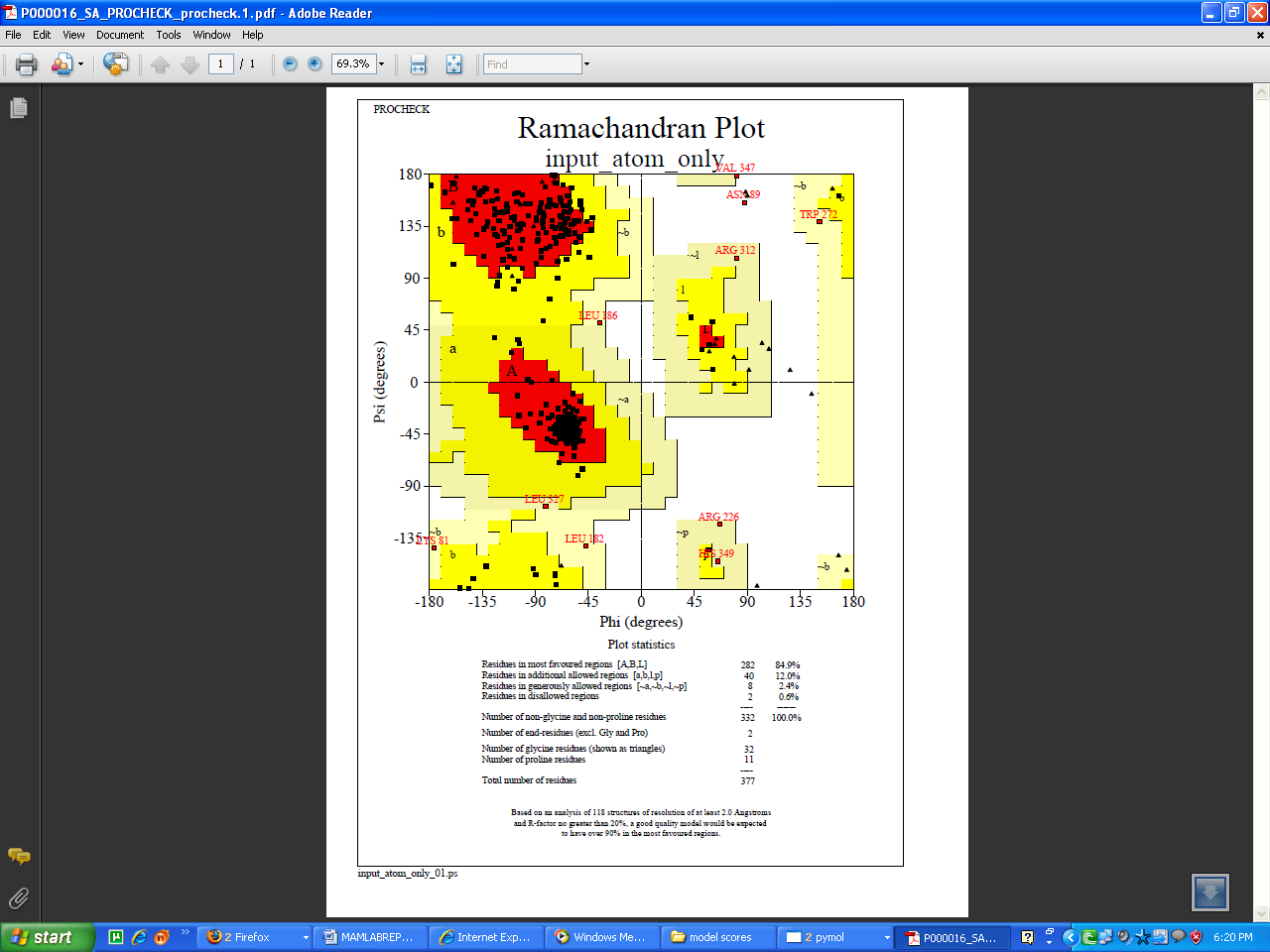 | Figure 3. Ramachandran plot of Homocitrate synthase built using Modeller9V5 |
3.2. Active Site Prediction
- Among the ten binding sites obtained from the Pocket finder (Fig 2B), the highly conserved site was identified using Q-Site finder. The binding pocket containing 26 residues such as ASN7, ASP8, THR9, ARG12, ASP13, GLU41, LEU42, GLU43, ILE46, LEU70, TRP72, HIS92, ALA94, GLY 134, GLY135, GLU136, ASP137, ARG160, ARG162, CYS164, THR 166, HIS193, HIS195, ASN217, THR218 and ASN229 were conserved and therefore considered for docking analysis.
3.3. Docking of Homocitrate Synthase with Potential Inhibitors
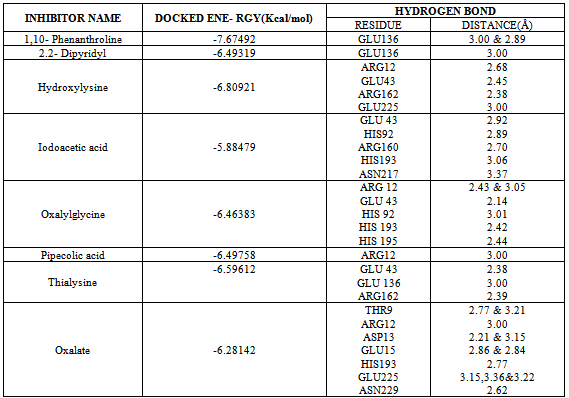
- Eight final docked conformations obtained for the different inhibitors were evaluated individually based on the number of hydrogen bonds formed; bond distance between atomic co-ordinates of the active site and inhibitor and the binding affinity (Fig 4). The docking energy showed that the inhibitors possessed the best binding affinity to the protein. Although the active site constituted about 26 residues, the ligands interacted with only 14 residues at THR9, ARG12, ASP13, GLU43, HIS92, GLU136, GLU225, ARG160, ARG162, HIS193, HIS195, ASN217, GLU225 and ASN229 respectively. However, among the chosen inhibitors, the best dock was performed by 1,10-Phenanthroline, which showed the energy of -7.67492Kcal/mol with two hydrogen bond interactions to the residue GLU at position 136 (Fig 4A) and Oxalylglycine which formed 6 hydrogen bonds (Fig 4E) but with slightly higher binding energy(-6.46383 Kcal/mol), than 1,10-Phenanthroline, with good bond distances. Iodoacetic acid and Oxalate may have least inhibition on protein due to the higher binding values when compared to other ligands and perhaps also showed more number of hydrogen bonds such as 5 and 12 respectively with higher bond lengths (Fig 4D and 4H). The hydrogen bond interactions between the eight inhibitors and Homocitrate synthase along with their bond distances and binding affinities are shown in Table 2.
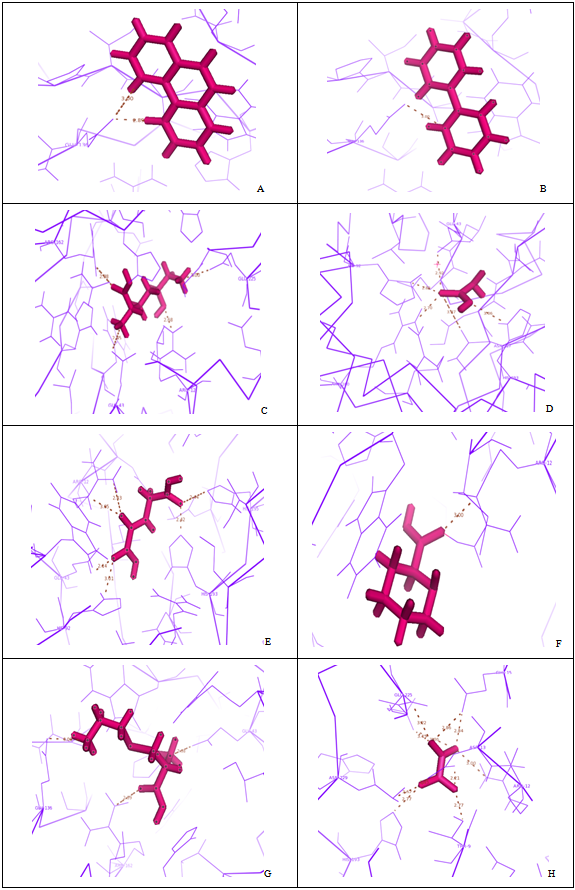 | Figure 4. Docking of ligands with homocitrate synthase |
|
- Proteins are shown in lines, ligand in stick mode and hydrogen bond as dotted lines. -A. Homocitrate synthase in complex with 1,10-Phenanthroline, B. Homocitrate synthase in complex with 2,2’- dipyridyl, C. Homocitrate synthase in complex with Hydroxylysine, D. Homocitrate synthase in complex with Iodoacetic acid, E. Homocitrate synthase in complex with Oxalylglycine, F. Homocitrate synthase in complex with Pipecolic acid, G. Homocitrate synthase in complex with Thialysine, H. Homocitrate synthase in complex with Oxalate.
4. Discussions
- The present study demonstrates the presence of genes for Homocitrate synthase and absence of genes for uptake hydrogenase in Nostoc punctiforme ATCC29133. Hence in this condition, Nostoc sp. produces hydrogen in excess than the other species having the genes for uptake hydrogenase. However, in spite of the production of enhanced hydrogen, the combined nitrogen atmosphere created by the active nitrogenase leads to the decrease in the net hydrogen production[5] reported that disruption of homocitrate synthase gene could lead to decrease in nitrogen fixation activity and increase in hydrogen production activity in Klebsiella pneumoniae. Homocitrate synthase produces homocitrate that binds to the FeMo cofactor of the nitrogenase, which is required for the efficient nitrogen fixation. Similarly, also in Nostoc sp., Homocitrate synthase functions in the biosynthesis of homocitrate that bind with the FeMo cofactor of nitrogenase but not with lysine synthesis through the alpha–aminoadipic acid pathway as reported by the same co-workers[5]. In this way, homocitrate synthase, a Zn containing metalloenzyme that catalyses the condensation of acetyl CoA and alpha-ketoglutarate to yield homocitrate and CoA[18] if inhibited could probably improve the chances of enhancing the hydrogen production. Hence, the activity of Homocitrate synthase was inhibited by metal-chelating reagents (1,10-phenanthroline, Oxalylglycine, Oxalate and 2,2’-Dipyridyl) and sulfhydryl binding reagent, Iodoacetic acid. In some of the yeast species, Homocitrate synthase is also believed to involve in biosynthetic pathway of lysine, in which Homocitrate synthase is inhibited by the lysine through feedback mechanism[21]. Since, some of the lysine analogues viz., Hydroxylysine and Thialysine have the complete inhibitory action on homocitrate synthase and in particular on alpha-ketoglutarate binding sites and that certain effector molecule viz., pipecolic acid also affected the co-operativity of S-acetyl coenzyme A binding sites[21], they were used as small molecules to examine their interaction on the active site of Homocitrate synthase.On analyzing the results obtained from the docking of metal chelating agents, was revealed to exhibit two hydrogen bonds and only one hydrogen bond respectively by 1,10-phenanthroline and Dipyridyl respectively. But in the case of Oxalate, it formed 12 hydrogen bonds in which some of the bonds showed higher bond length (more than 3Å) that lead to the weaker interactions. In the case of Sulfhydryl binding agent Iodoacetic acid also formed more number of hydrogen bonds (five) like Oxalate, which exhibited the least effect on protein due to its higher docking energy[22]. In the case of Lysine analogues, Hydroxylysine and Thialysine formed four and three hydrogen bonds respectively, but at the same time had a good inhibition for protein. The affector, Pipecolic acid formed only one hydrogen bond with the binding energy of -6.49758 Kcal/mol, which in spite of good binding energy, could not be considered as best because of least inhibitory action on protein due to its highest docking energy and the least number of hydrogen bond formed. However, based on the comparison of the analysis and with relative to binding energies, 1,10-Phenanthroline could be identified as the best potent ligand with very least binding energy of -7.67492Kcal/mol than other inhibitors. Since Homocitrate synthase is a Zn-containing metalloenzyme, Zn metal is supposed to bind to the amino acids such as cysteine, Histidine, Aspartic acid, Glutamic acid, Asparagine, and Glutamine (given according to priority)[23-24]. Hence according to the least binding energy and the amino acids binding to the zinc metal, 1,10-Phenanthroline was revealed to be an effective interactor of the protein Homocitrate synthase. But according to the number of hydrogen bonds formed and the length of the hydrogen bonds and the amino acids binding to the zinc metal, Oxalylglycine is considered to be the best since it formed six hydrogen bonds of which four were bound to the appropriate aminoacids such as GLU43, HIS92, HIS193 and HIS195 but with slightly higher energy (-6.46383 Kcal/mol) than 1, 10-Phenanthroline with good bond lengths (less than three).
5. Conclusions
- Since Homocitrate synthase is a Zn-containing metalloenzyme, it was shown to be effectively inhibited by the metal chelating agents, of which 1,10-Phenanthroline and Oxalylglycine showed the higher effect of inhibition when compared to other inhibitors under investigation. Both 1,10-Phenanthroline and Oxalylglycine showed very good binding energy and interactions with the active site residues compared to the other inhibitors. But basically any insilico analysis is either partial or incomplete if unproved under experimental (in vitro) conditions. Hence, in the direction of understanding the inhibitory mechanism, the work should be taken up to be implemented in wet lab which is our fore-front research (proposal applied to funding agency).
Webography
- http://genome.kazusa.or.jp/cyanobase/NPUNhttp://www.uniprot.org/uniprot/B2J701http://www.rcsb.org/http://bmm.cancerresearchuk.org/~3djigsaw/http://www.salilab.org/modeller/http://expasy.org/spdbv/http://nihserver.mbi.ucla.edu/saves/http://www.modelling.leeds.ac.uk/qsitefinder/http://www.ncbi.nlm.nih.gov/Structure/index.shtmlhttp://www.arguslab.com/downloads.htm
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-Text HTML
Full-Text HTML