-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Advances in Life Sciences
p-ISSN: 2163-1387 e-ISSN: 2163-1395
2018; 8(2): 39-50
doi:10.5923/j.als.20180802.01

Osmotin: A PR Gene Imparts Tolerance to Excess Salt in indica Rice
Asit B. Mandal1, 2, Anusrita Biswas1, Pranit Mukherjee2, Raju Mondal2, Sourav Dutta2, Chiranjib Mandal2
1Central Agricultural Research Institute, Port Blair, India
2ICAR-Central Research Institute for Jute and Allied Fibres, Barrackpore, India
Correspondence to: Asit B. Mandal, Central Agricultural Research Institute, Port Blair, India.
| Email: |  |
Copyright © 2018 The Author(s). Published by Scientific & Academic Publishing.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Agrobacterium-mediated genetic transformation (strain: GV2260 harbouring osmotin gene in a binary vector pGV2260 under the transcription control of CaMV 35S promoter and Nos terminator) was restored to develop productive salt tolerant transgenic rice varieties in a well adaptive high yielding indica rice background of var. Ananda. Out of 705 calli transformed, 426 survived (60.44%) on kanamycin (50 mg/l) supplemented MS medium, which could regenerate 78 plantlets of which 56 survived from 29 independent transformation events. Molecular analyses showed 23 PCR positive, 9 Southern positive and 6 plants positive to both PCR and Southern hybridization. Southern hybridization with radio-labeled osmotin probe showed transgene integration in 2-6 loci in 6 representative transgenic plants. Seed germination test involving seeds at T1 generation on MS medium containing 50mg/l kanamycin showed 3:1 ratio for majority of the lines albeit with a few exception, might be due to small sample size or epistatic interactions. Tolerance test to excess salt was conducted invivoon artificially stimulated NaCl stress under hydroponics involving Hoagland solution showed progenies of T46-1 were most tolerant to excess NaCl followed by the progenies of T2-6, which were planted on soil in cement pots in the Transgenic Rearing Facility (Category II, DBT approved) till maturity and harvested seeds were kept plant wise for subsequent generation advancement of most productive lines with desirable salt tolerance for cultivation enmasseunder saline soil for enhanced productivity under constrained soil at sustainable-scale in Andaman Islands and elsewhere.
Keywords: Osmotin,Agrobacterium-mediated genetic transformation, Indica rice, Salt tolerance, Andamans
Cite this paper: Asit B. Mandal, Anusrita Biswas, Pranit Mukherjee, Raju Mondal, Sourav Dutta, Chiranjib Mandal, Osmotin: A PR Gene Imparts Tolerance to Excess Salt in indica Rice, Advances in Life Sciences, Vol. 8 No. 2, 2018, pp. 39-50. doi: 10.5923/j.als.20180802.01.
Article Outline
1. Introduction
- Rice is one of the most important cereal crops, providing staple food for nearly 40% of the global population. More than 91% of rice is grown and consumed in its homeland Asia (Khush & Toenniesen, 1991, Khush & Baenziger, 1996), where it has been originated and domesticated. Indica rice varieties alone feed about 3 billion people in the developing countries (Datta, 2002). To meet increasing demand of additional rice to feed the booming up population, production has to be increased at least 60% by next 25 years (Khush & Baenziger, 1996). Conventional breeding has contributed moderately by developing large number of high yielding varieties (HYVs) and by releasing those in public domain for large-scale cultivation. However, the pace of progress is highly time consuming, tedious and often misfires. To ensure crop genetic improvement preciously with ease and confidence, modern plant biotechnological tools and techniques as available since mid-eighties, have substantially enhanced productivity and production in many countries globally. Now-a-days rice is considered to be a model system in cereals for cell technology works especially in developing transgenic lines involving different genes from heterologous sources across the world. Virtually it has erased the transsexual barrier among the biological organisms. Thus made the entire living world a universal source of useful gene/s for use in genetic improvement programs even for those traits, which were considered to be unamenable for modification through classical plant breeding. This has encouraged us to undertake the challenging job of development of salt tolerant high yielding indica rice variety by dove-tailing an unique gene- osmotin, which possess proven track record to impart tolerance to both biotic and abiotic stresses. Salinity is a serious abiotic stress limiting agricultural productivity. In India out of 44 million hectares of land where rice is grown about 30% (~13 million hectares) contain excess salt and 10 millon hectares remain fallow (Annual Report 2015-16, http://agricoop.nic.in/). About one third of irrigated land available globally is salty due to secondary salinization (Epstein et al., 1980) globally, which is estimated to be 50% of cultivable land by 2050 is a serious concern (Sarangi et al., 2011). In Andaman & Nicobar islands (a constituent union territory of Indian Republic), where rice constitutes the lifeline crop for sustenance and perpetuation of 4.5 lakh people. Rice is principally cultivated in these islands on 12,000 ha low lying areas. Against the domestic demand of 64,000 tons, the islands are currently producing ~31,650 tons with a chronic production-requirement gap of almost 50%, which is being met by transporting additional grains from mainland India (which is very tedious, risky and quarantine offensive). In addition, about 4,000 ha of saline soil is available containing different divalent cations like chlorides, sulphates, carbonates and bi-carbonates, in the coastal belt majority of which remain idle and offers an alternative for rice husbandry. Soil of Andaman’s’ as such is acidic in nature and often gets inundated with estuarine water during tidal flash. A tall, traditional, highly photosensitive Burmese (Presently Myanmar) rice cultivar of extremely long duration (~180 days) viz. C14-8 is cultivated on large-scale and is very popular among the islanders with average yield of ~2.6 tons/ha, however, in the saline soil C14-8 yields only about 1 ton/ha (Mandal et al., 2004). A few other traditional cultivars viz. Nata, Mahsuri, Nona etc. are also cultivated with about 1.5 tons/ha grain yield. Till date, India has released more than 40 salt tolerant rice varieties among, which IR 21820-154-3-2-2-3 and MTU 76339 (Swarna) were found to produce about 2 quintal/ha if the rain fall remains 3000-3300 mm and distributed during the entire rice growing season in Andamans. At the interface of global climate change, the agricultural productivity is declining in major food crops (Varshney et al., 2011). Rice is a sensitive crop to drought and salinity stress (Flowers & Yeo, 1981, Yeo & Flowers, 1984, Khush & Tonniessen, 1991, Khush & Baenzier, 1996) and highly sensitive to submergence stress (Hossain, 1996). To circumvent this problem one strategy is to develop plants tolerant to salinity stress. For example, in tomato plants tolerant to 150 mM NaCl has been developed (Goel et al., 2010). The present investigation was conceived to harness the benefit of osmotin gene in developing salinity tolerant (especially to excess NaCl) indica transgenic rice varieties to enhance rice productivity on saline soils through large- scale cultivation.Plant responses to abiotic stresses largely depend upon cascade of events involving stress perception, signal transduction and expression of stress related specific biomolecules (Vincour and Altman, 2005) conferring stress tolerance or enzymes present in pathways led to synthesis of structural metabolites and osmolytes (Wang et al., 2003; Seki et al., 2003; Shinozaki et al., 2003; Rontein et al., 2002). One such gene - osmotin (belongs to PR5 protein family (Yun et al., 1998; Lee et al., 2010; Singh et al., 1989) encodes osmotin, a stress-responsive multifunctional 24-kDa protein, that is induced in response to salinity and drought, which was originally isolated from tobacco and used extensively in 1980s (Singh et al., 1989, b; LaRosa et al., 1992) in imparting abiotic stress tolerance by providing osmo-tolerance to plants. Osmotin also acts as an antifungal cytotoxin that causes rapid cell death in yeast (Saccharomyces cerevisiae) and exhibits cryoprotective functions (Kupchak et al., 2008). In olive tree, osmotin provides cold protection via different mechanisms related with programmed cell death (D’Angeli & Altamura, 2007). Jami et al., (2007) reported that Solanum nigram osmotin-like protein (SniOLP) is a PR protein and possess antifungal activity. The SniOLPprotein encoding a gene from Solanum nigram represents a small multigene family and displays organ-specific expression (Jami et al., 2007). Similarly, in strawberry, presence of osmotin-like protein gene (faolp2) belongs to a multi gene family has been reported (Zhang and Shih, 2007). Since then, physiological role of osmotin in abiotic and biotic stress tolerance management studied extensively (Kononoweiz et al., 1992; Jami et al., 2007; Kupchak et al., 2008; Lee et al., 2010), although exact mechanism in diverse situations are not clear. This work uses a simple approach to introduce a heterologous alien transgene through Agrobacterium- mediated genetic transformation in to genome of a popular well adaptive high yielding indica rice variety for imparting tolerance to excess salt. Abiotic stress of different types can affect almost all field crops grown in natura. Further, over-expression of the gene in transgenic plants is known to impart tolerance against various biotic and abiotic stresses including cold, excess salt and drought (Goyary 2009; Parkhi et al., 2009; Goel et al., 2010; Sarangi et al., 2011). Although there are some reports available on the plausible mechanism through, which osmotin imparts defense against biotic stresses, its role in abiotic stress response is still unclear. It is also reported that high proline accumulation in osmotin over- expressing transgenic plants in response to cold, salt or drought stressess (Parkhi et al., 2009; Goel et al., 2010; Barthakur et al., 2001), suggesting existence of cross-talk between different stress response pathways. Thus, it appears that osmotin gene can act through mediation of increased proline content. Enhanced proline content under abiotic stress condition has been found to be positively correlated with increased activity of ɣ-pyrrolin-5- carboxylate reductase (Madan et al., 1995) and y-glutamine kinase (Girija et al., 2002) and with low activity of proline oxidase and proline dehydrogenase, both are functionally degrading enzyme. ɣ-pyrroline-5- carboxylate reductase enzyme catalyses proline synthesis from glutamate. Overexpression of this gene reduces tolerance to abiotic stress through accumulation of proline and displayed increased plant biomass; grain yield etc. under salinity and drought stress conditions (Kavi et al., 1995). Accumulation of proline in transgenic lines with over expression of osmotin gene in tobacco has been reported (Barthakur et al., 2001, Sokhansanj et al., 2006). Transgenic plants those, which are able to produce proline, behaves as normal plant, however, failed to display tolerances to biotic and abiotic stresses (Nanjo et al, 1999 & Xin et al, 1998). Osmotin has been found to interact with other proteins to activate signal transduction pathway leading to defenses gene expression by changing the balance of cell death and protein survival in the plant system. This pathway showed interaction of at least two independent cell death pathways activated either by NaCl or abscisic acid (ABA).This study presents the results of an elaborate experiment to integrate and express of osmotin gene in an indica rice var. Annada through Agrobacterium-mediated genetic transformation as the recipient system.
2. Materials and Method
2.1. Plant Materials
- One high yielding well adaptive indica rice variety - Annada was obtained from Directorate of Rice Research (ICAR-DRR; Presently ICAR-Indian Institute of Rice Research), Hyderabad. Mature healthy seeds were dehusked manually and immersed in 3% (v/v) aqueous Teepol solution (anionic detergent, Sigma-Aldrich) with continuous shaking for 10 min at 80 rpm on a rotary shaker (Remi). The seeds were thoroughly washed and surface sterilized with freshly prepared 0.02% HgCl2 (E.Merck) under laminar air flow (LAF) for 10 min followed by wash with sterile double distilled water three times. The surface sterilized seeds were blot dried with autoclaved tissue paper. 200 axenic seeds were cultured on MS-callus induction medium (CIM) solidified with 0.8% agar (Hi-Media) pH 5.8 in rimless culture tubes (Borosil make, 22mm dia) and closed with non-absorbent cotton plugs (Murashige and Skoog, 1962). The culture tubes with surface sterilized single seed were kept in dark at 25±2°C. After ten days, loose calli started developing at the swollen junction of radicle and mesocotyl. Ten days old calli were excised out from scutellar surface of the germinating seedlings and placed on the callus maintenance medium (CMM) as mentioned in Table 1. Calli from individual seeds were maintained individually and sub-cultured at 21 days interval for four times till very friable embryogenic calli were found to develop for undertaking genetic transformation work.
2.2. Bacterial Strain, Plasmid and Culture Condition
- Transformation experiments were conducted using Agrobacterium tumifaciens strain GV2260 harboring pGV2260 binary vector. T-DNA region of this binary vector had nptII (neomycin phospho-transferase) gene for kanamycin resistance (as plant selection marker) for selection of the regenerates containing the desired gene- osmotin (0.75kb), which was ligated at BamHI/XbaI site under the transcription control of a strong constitutive CaMV35s promoter and CaMV35s terminator (Figure 1). Agrobacterium cultures were maintained on LB medium following Sambrook et al. 1989 supplemented with 100mg/l rifampicin (Sigma-Aldrich) 50mg/l kanamycin (Sigma-Aldrich) and 100mg/l carbenicillin (Sigma-Aldrich) and solidified with 15 g/l agar (Hi Media). A single bacterial colony was transferred to liquid LB medium containing same antibiotics contents and grown for 48 h at 28°C at 220 rpm in a bacteriological BOD incubator shaker (Lab-line). The bacterial cells were pelleted in a tabletop centrifuge (Remi) at 4,000 rpm at ambient temperature for 10 min, followed by re-suspending the pellet in Agrobacterium culture medium (Table 1) to a final OD600 of 0.5 for vir gene induction. Finally, the Agrobacterial cells were incubated and kept at 28°C for 5 h before undertaking genetic transformation work involving highly embryogenic actively growing calli from Annada.
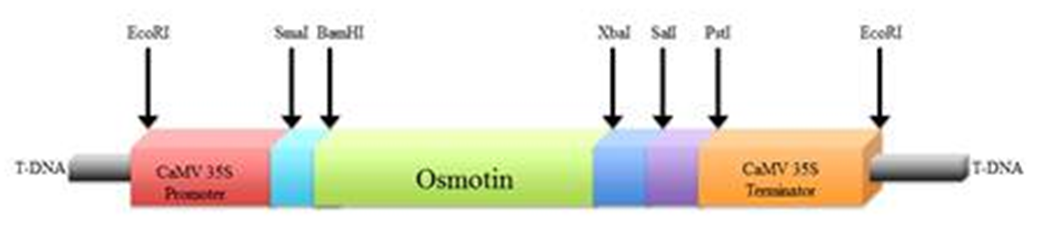 | Figure 1. Partial physical map of pGV2260 harbouring osmotin (0.75kb) under the transcription control of CaMV35S promoter (0.8kb) and CaMV35S terminator (0.2kb) |
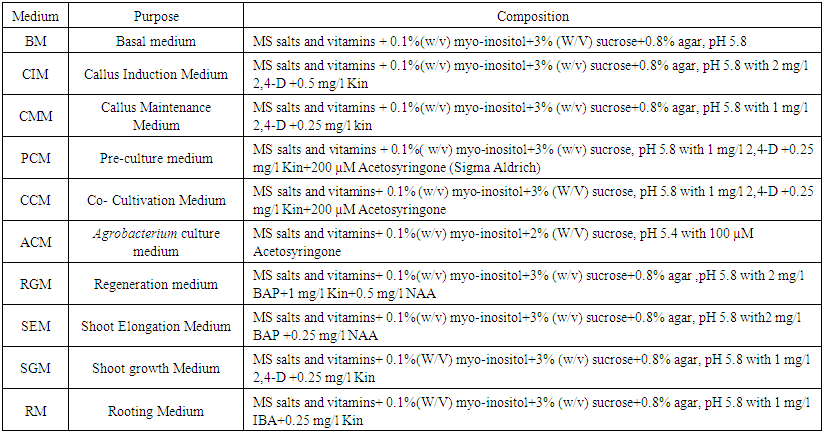 | Table 1. Different media composition used in in vitro tissue culture and Agrobacterium-mediated genetic transformation of indica rice var. Annada |
2.3. Agrobacterium-Mediated Genetic Transformation
- Ten days old calli were found to be most suitable to pre-culture in pre-culture medium prior to Agro-infection. Pre-cultured calli were punctured at 2-3 points randomly with a sterile needle (needle diameter ~ 0.5mm). Wounded calli were subsequently immersed in the pre-cultured Agrobacterium suspension (in ACM medium for 20 min, Table 1) followed by vacuum infiltration under the pressure of 600mm Hg for 10 min. Inoculated calli were blotted dry on sterile filter paper to remove excess Agrobacterial cells and co-cultured for 72h in radio-sterile petridishes (Tarsons, 90mm dia) containing Whatman filter paper (1 mm thick) spread and moistened with co-cultivation medium at 25±2°C.
2.4. Stable Transformation, Selection and Plantlet Regeneration
- After 72h of co-cultivation in dark at 26±1°C, the co-cultured calli were washed with sterile distilled water supplemented with 250mg/l cefotaxime (Sigma-Aldrich). After 2-3 times wash, the calli were blotted dry on sterile filter paper and cultured on selection medium (MS medium) supplemented with 75ml/l kanamycin. After two rounds of selection on CMM (Table 1), survivor calli were transferred to regeneration medium (RM, Table 1). Regenerated plantlets were further transferred on to MS medium supplemented with 50mg/l kanamycin and incubated under 16/8 h light (130 μE /m2 /s) dark for 7 days. When the plantlets attained a height of 4-6 cm, those were transferred to rooting medium (RM, Table 1) for inductionof roots. The rooted plantlets were transferred on to culture tubes containing Hoagland solution for acclimatization for a week and subsequently shifted to Transgenic Rearing Facility (Category-II, DBT approved) where those well-acclimatized plantlets were grown in autoclaved wet soils in small plastic pots. At active vegetative phase, the plantlets were shifted to soil in big cement pots (1x1x1.5feet) in the transgenic house. During anthesis, individual panicles were enclosed with butter paper envelopes to prevent pollen escape and subsequently to avert cross-pollination. Those plants were grown to maturity under fine mesh net cover. Seeds were collected, threshed, dried and kept separately plant wise for raising the progenies in the succeeding generations.
2.5. Molecular Analysis
2.5.1. DNA Isolation
- Genomic DNA was isolated following CTAB method (Murray and Thompson, 1980) from tender leaves of one-month-old putative transgenic plants (primary regenerants, To generation) and purified using RNase (Amersham Pharmacia Biotech Inc., USA). The purified genomic DNA was quantified through both spectrophometrically (UV-Vis spectrophotometer, Chemito) as well as through co-electrophoresis using a horizontal electrophoresis apparatus (Bio-Rad) along with λ marker (NEB), using TAE buffer, pH8.0 at 100V in ethidium bromide (EtBr) mixed 0.8% agarose gel till the tracking dye reach to the opposite end of the lanes loaded with genomic DNA. Average of both was taken in calculating actual DNA concentration of samples to undertake other molecular analyses to confirm the transgenic status of the putative transgenic plants.
2.5.2. PCR Analysis
- PCR provides very simple and efficient tool in detecting transgenic status and is conventionally used to assess the presence of transgene in the primary regenerants. To amplify osmotin (0.75kb), forward primer 5’GAAGTGACTCAATTTTA3’ and reverse primer 5’CCTGGCCGAGTACGCCTT3’ were used, whereas nptII was amplified using forward primer 5’TCGGGAGCGGCGATACCGTA3’ and reverse primers 5’GAGGCTATTCGGCTATGCTG-3’. In all PCR reactions, 100ng of plant DNA and negative control (untransformed plant DNA) and 20-25ng positive control (pGV2260) DNA were used in a total reaction volume of 25µl [containing 1X Taq reaction buffer (10X), 200 µM dNTPs (10mM), 1.25 units/50 µl PCR Taq DNA polymerase, 0.2 µM primers (10 µM forward and 10 µM reverse primer) in nuclease free water] was performed using a thermal cycler (Bio-Rad, USA) by adopting the programme: one cycle at 94°C for 1 min, 35 cycles of 94°C for 1 min (denaturation), 40°C for 2 min (annealing temperature for nptII) or 57°C for 1 min (annealing temperature for osmotin) and 72°C for 3 min (osmotin extension) and 5 min (nptII extension). The amplified products were electrophoresed on 0.8% agarose gel containing ethidium bromide (EtBr) in TAE buffer (pH 8.0) and photographed in a gel documentation system (Bio-Rad, xr+ gel documentation system).
2.5.3. Southern Hybridization
- PCR positive and kanamycin resistant plantlets developed from transformed calli of var. Annada indicated their transgenic status tentatively. However, to confirm transgenic status stringently further, genomic DNA from transgenic plantlets was used for Southern blot hybridization to detect the presence of osmotin gene following standard protocol (Sambrook et al., 1989). With this target, 2 μg of genomic DNA was digested with PstI (NEB). Resulting fragments were size fractioned on 1% TAE agarose gel and subsequently DNA was denatured in situ under alkaline condition followed by neutralization. The restricted fragments were transferred from the gel to a Hybond-N+ nylon membrane (Amersham) with the help of an instrument [a gift from Centre for Cellular and Molecular Biology (CCMB), Hyderabad] using neutral buffer. The membranes were cross-linked at 1200 J for 2 min. Probe for osmotin fragment was prepared upon digestion of pGV2260 measuring 0.75 kb by using restriction enzymes BamHI/XbaI. 25 ng DNA was denatured by heating in a boiling precision water bath (ThermoScientific) at ~100°C for 5 min and the denatured DNA was added to the labeling mixture (α32 P dCTP procured from Jonaki, BRIT, Hyderabad, a supply outlet of BARC, Trombay, Mumbai) and mixed well. Redi-prime DNA labeling System (Amersham Life Science RPN) was used for radio labeling of the probe. The mixture was added and mixed by repeated pipetting up and down for 5 times followed by centrifugation and then incubated at 37°C for 10 min followed by snap chilling on ice. Reaction was terminated by addition of 5µl 0.2mM Na-EDTA. Radio labelled probe was added to the hybridization buffer (30ml) in a hybridization bag made up of thick polyethylene and sealed perfectly to avert any leakage. The probe was added to the hybridization solution carefully and was kept at 65°C (overnight) with gentle shaking. Pre-hybridization and hybridization were done at 65°C (1h), with addition of α32 P dCTP (radio-labelled probe) and salmon sperm DNA (Sigma-Aldrich) to the hybridization solution, which was again kept at 65°C (overnight) in a shaking water bath. Following which, the nylon membrane was washed with the washing solution, dried and covered with Saran wrap. After stringent washes the membranes were then exposed to X-ray film (Kodak) with intensifying screen at -86ºC in 8”x10” HyperTM cassette. Southern blot showed the band indicating presence of osmotin in some putative transgenic plants (PCR positive plants) but not in all. If the transgene is integrated in the putative transgenic plants, signal sensed from the blot are expected to be developed in the x-ray film used for the Southern hybridization.
2.6. Inheritance of Transgene Based on Seed Germination Test
- Seeds from 9 transgenic plants (T0 generation) were harvested after maturity plant wise, threshed cleaned and kept individually. Those seeds were used for seed germination test. Twenty-five seeds were dehusked manually, cleaned and surface sterilized with aqueous Teepol solution (5%) and washed thrice. Those were further immersed in freshly prepared 0.1% HgCl2 (E.Merck) for 10 minutes and subsequently washed thoroughly with autoclaved dH2O four times under laminar air flow (LAF) bench. For germination of seed, 25 axenic seeds were cultured on half strength MS medium supplemented with 50µg/ml kanamycin and kept in dark at 25±2°C in the culture room. After 3-4 days, number of germinated seeds was recorded. After 15 days survivor seedlings were grown in soil in the Transgenic Rearing Facility. In this stage, those were screened again through PCR, using the method described previously. Seed obtained from the T1 plants were germinated under kanamycin antibiotic selection pressure (50 µg/ml) to assess the expression of osmotin in T2 generation. Furthermore, seeds from T1 plants were also tested for germination on the medium supplemented with 50 µg/ml of kanamycin along with untransformed control.
2.7. Salt Tolerance Assay: in vivo Assessment
- Mature seeds from T1 plants were harvested and were used to assess their tolerance norm under excess NaCl in a hydroponics culture. Seeds were germinated in rows (channels were made up of Styrofoam sheet (1.5 inch thick) with mosquito net stitch underneath. The entire sheets with 10 rows of germinated seeds in the each lane were floated under hydroponics in Hoagland solution, pH 5.0 in a 4l capacity (16x12x3inch) plastic tray. The under surface of the tray was painted black with enamel to prevent the illumination, which may reduce the rooting response since root growth is reported to be negatively phototrophic. When the seedlings attained 2-3 leaf stages, salinity stress to tune of 8dSm-1 was created through application of NaCl (SRL). Pokkali and IR 28 varieties were used as salt tolerant and susceptible checks, respectively. Seeds of var. Annada were used as parental control check. After 28 days, only the survivor plants were shifted to saline soil in cemented tank under artificially simulated wet soil control condition fortified with NaCl. During culture, water level was maintained at 3-4 mm and the pH was maintained at 5.0 with the aid of KOH or HCl solutions. NaCl tolerance norms to excess salt were scored in 0-9 scale (SES 1988, IRTP, IRRI, Philippines 1984) towards salinity maintained at 8 dS/m during the remaining life span of the crop till maturity. Major agronomic characters and tolerance levels towards excess NaCl were measured/scored and data were analyzed using IRRISTAT software (IRRI, Philippines).
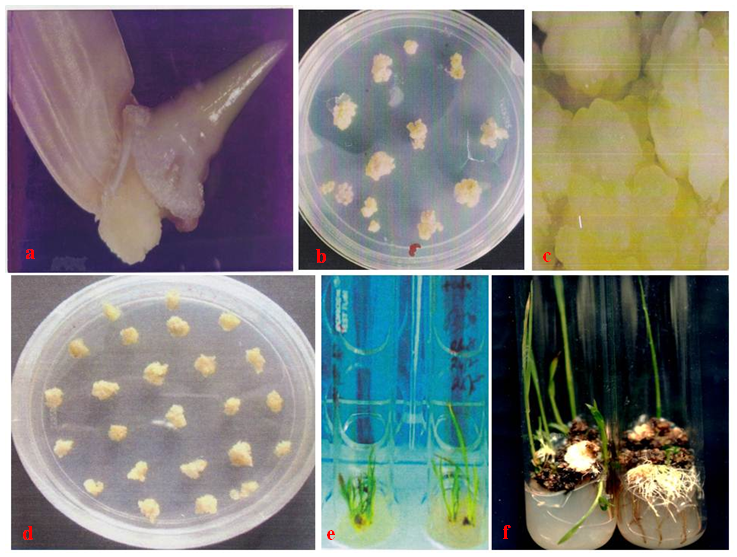
3. Result
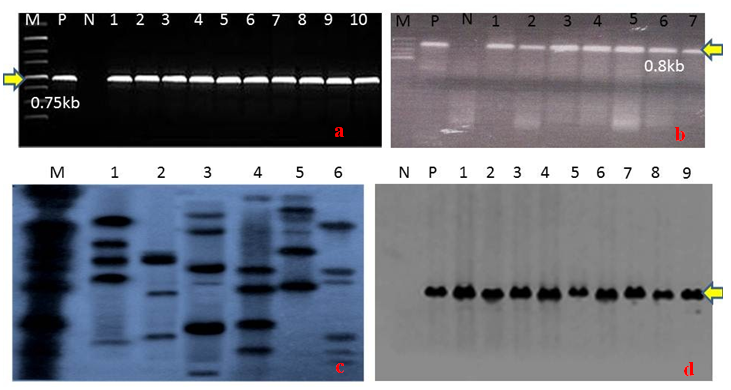
- Embryogenic calli were induced profusely on MS medium containing 2mg/l 2,4-D. During transformation most of the calli died on kanamycin supplemented selection medium. In R1 replication, 113 calli (64.45%) out of total 175 calli survived (Table 2). The survivor calli were allowed to differentiate in presence of kanamycin (50mg/l) and 55 calli were regenerated into 21plantlets. High concentrations of BAP (≥ 2 mg/l) and Kin (≥1mg/l) in the regeneration medium were found to facilitate enhanced plantlet regeneration. After initiation, the plantlets were allowed to grow to the height of 4-6 cm when those were transferred onto shoot elongation medium (SEM) (Table 1) containing MS with 2mg/l BAP. Most of the regenerants were found to be slender and those were allowed to grow for next 15 days on the same medium. Shootlets were shifted onto root induction medium containing MS with 1 mg/l IBA (Figure 2.). After 15 days, the plantlets with profuse roots were acclimatized in Hoagland solution (Yoshida et al 1976) for a week time and then transferred for hardening to wet sterile soil filled plastic pots in transgenic house. At active tillering phase the plantlets were further transferred on to big cement pots (1x1x1.5feet) till maturity (Figure 3a. & 3b.). Similarly in case of R2, R3 and R4 replications, 105 (56.75%), 102 (60.00%) and 106 (60.57%) calli survived out of 185, 170, 175 calli, respectively. Across four replications, an average of 60.44% calli was found to survive and 51.01% calli showed plantlet regeneration and 49.52% plantlets developed from regenerative embryogenic calli. Many plantlets become albino due to bleaching action of kanamycin, which was reported by many investigators in a wide range of experiments involving diverse plants (Biswas, 2002). Through histological observations, plantlets were found to be regenerating following both somatic embryogenesis and organogenetic pathways (data and figures not presented). Survival percentages of putative transgenic plants were found to be very low, only 52 plantlets finally survived from. Among 29 independent transformation events, 78 plantlets were regenerated of which 56 (71.79%) plantlets survived (Table 3). However, during the course of acclimatization, hardening, in vivo assay for assessment of tolerance norms under hydroponics and at different stages of growth till maturity, altogether 31 plants died or did not set fully mature filled grains. The remaining vigorously grown and apparently looking normal putative transgenic plants were analyzed at molecular level in respect of PCR, Southern blot hybridization, physiological assays and transgene inheritance studies involving T0 and the seeds at T1 generation. Out of 56 plantlets, 23 (41.07%) showed PCR positive result. Result demonstrates the presence of the band of expected size in PCR positive (compatible primers for both osmotin and nptII were used) transgenic plants (0.70kb) (Figure 4a.) and for the probe (0.8kb) (Figure4b.) where as control plantlet did not show any fragment amplification. This result clearly indicates true transgenic nature of the putative transgenic plants tested. Southern blot also revealed integration of the transgene-osmotin in all PCR positive plants as mentioned above and no rearrangements were observed in any of the transgenic plants studied. Southern blot analysis indicates the stable integration pattern of the transgene in the genome. PCR and kanamycin resistant nature of the plantlets tentatively advocates their transgenic character. Anyhow to reconfirm the transgenic status more stringently, extracted genomic DNA were digested with unique restriction enzyme PstI having single cut site outside the coding sequence of osmotin. This could digest the rice genomic DNA in two different fragments of random length limited by PstI site. Based on the presence of transgene at different integration sites in the nuclear genome, radiolabeled osmotin probe was found to hybridize with genomic DNA, which was immobilized on the charged nylon membrane through Southern transfer. Among 56 survivor plantlets, 9 (16.07%) were found to display positive result in Southern blot hybridization (Figure 4c. & 4d.). A representative Southern blot involving 6 PCR positive putative transgenic plants along with DNA from a non-transformed control plant (Annada) is displayed. In essence, among 78 plantlets, only 56 (71.79%) survived, which were ultimately used for molecular analyses. Out of those 23 (41.07%) plantlets showed PCR positive results in respect of nptII gene and 9 plantlets (16.07%) showed positive result in respect of Southern hybridization with radiolabeled osmotin probe. The above findings indicate that Agrobacterium–mediated genetic transformation hold enormous prospects in indica rice genetic improvement through transgenic development precisely with ease and confidence. The transgenic plants derived from Annada were looking normal with no ectopic expression of any characters and those produced fully fertile seeds in the transgenic house. Seed germination test was performed involving seeds from transgenic plants of T0 generation on MS medium containing kanamycin (50 mg/l) involving 25 seeds from each plant (Figure 3c.). In most cases the proportion of resistant versus sensitive plants were found to be not significantly different from 3:1 ratio (P > 0.05) (Table 4), which indicates the presence of single dominant transgene gene in operation. Plants that did not follow 3:1 ratio, the distorted ratio might be due to use of extreme small size of the seed, interaction with the chemical environment in which the seeds were germinated or epistatic interactions. Seeds of T1 progenies showed differential survival percentage under salt stress (8 dS/m) (Figure3d.), which was found in the range of 38.23-88.00% (mean = 64.12%). Maximum and minimum survivals were observed from the seeds of T46-1 and T2-6 plants, respectively. In case of control set (untransformed Annada plants) survival percent under NaCl stress were found to be 28.0% (NaCl tolerance score was recorded at vegetative, active tillering, flowering and maturity phases). At vegetative phase the tolerance score was found to range in between 2-7. Maximum tolerance towards NaCl stress at this phase was observed in T23-2 and T46-1 (Tolerance score -2) and minimum tolerance in T2-6 and T25-7 (tolerance score -6). While considering tolerance values at active tillering phase T23-2 and T46-1 were found to display salt tolerance (tolerance score 2) In the flowering stage the aforesaid two plants showed average tolerance value of 2 and 3, respectively. In contrast, T2-6 showed minimum tolerance score (tolerance score 6) at maturity, T23-2 and T46-1 were found to be maximally tolerant (tolerant score -3), whereas T2-6 show minimal tolerance (tolerance score -7) towards excess of NaCl. Considering tolerance values across four phases, T46-1 was found to be maximum tolerant among all progenies tested (Table 5).
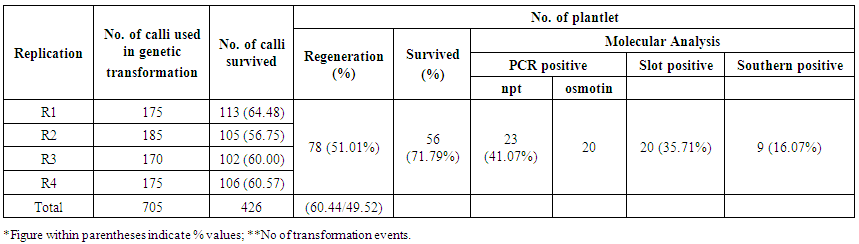 | Table 2. Synopsis of Agrobacterium-mediated genetic transformation in indica rice var. Annada |
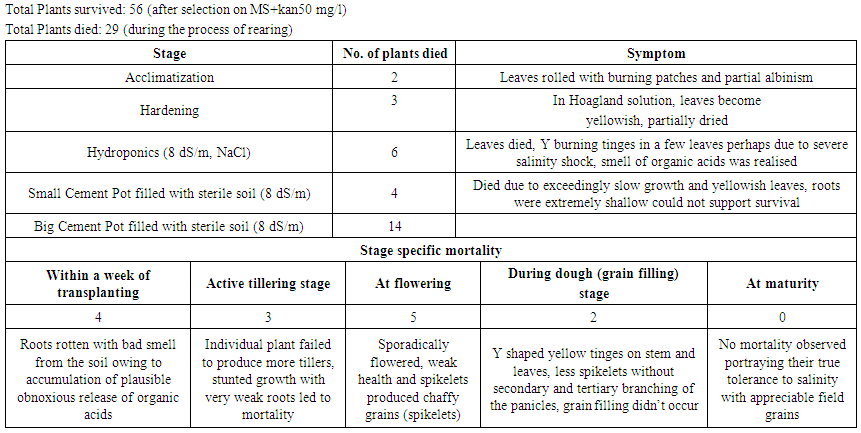 | Table 3. Mortality of the putative primary transgenic lines of Annada harbouring osmotin at different stages of rearing till maturity |
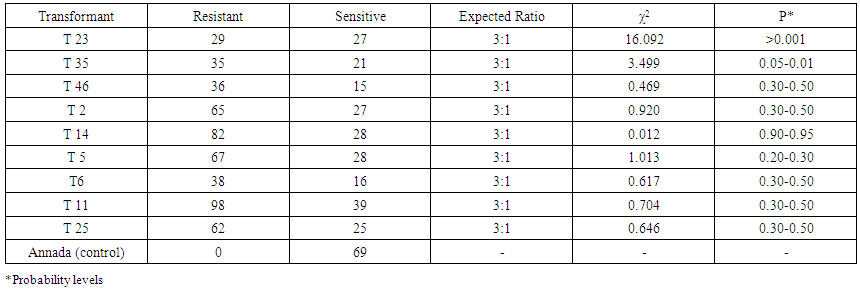 | Table 4. Germination test of seeds obtained from putative transgenic plants of var. Annada on medium containing 50 mg/l kanamycin at T1 generation |
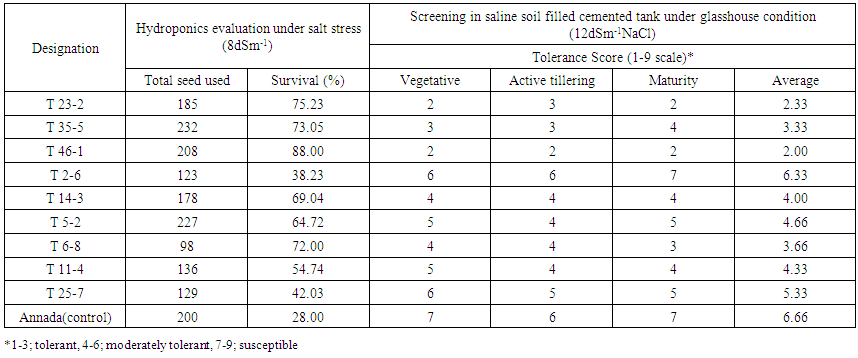 | Table 5. Performance of a few promising putative transgenic lines of var. Annada under in vivo assessment of tolerance towards excess NaCl involving seeds of T1 generation |
4. Discussion
- Osmotin gene is induced by different types of abiotic stresses such as salinity, temperature and drought etc. (Singh et al., 1989; LaRosa et al., 1992, Zhu, 1993). It was reported to impart tolerance to salinity stress in potato (Goel et al., 2010), rice (Vinod et al. 2013) and in many other crops (Apse & Blumwald, 2002; et al., 2005). Osmotin gene in different plants showed varied physiological responses but the intrinsic underlying mechanism still remains unclear in response to excess NaCl stress. In this study, plantlets were regenerated well by using in vitro culture technique and through standard transgenic development procedure, we introduced an osmotin gene in an important crop - indica rice and provided evidences that the transgenic plants can survive well under salinity stress. Survival of those plants was verified through salinity tolerance test under hydroponic culture system. Analysis of several molecular parameters such as PCR, Southern blot hybridization, physiological assays and transgene inheritance studies clearly demonstrated the development of osmotin endowed transgene line of indica rice.Modern biotechnology accelerated the development of genetically modified (GM) crops production. As a result cultivation area of GM crops increased more than 100-fold from 1.70 million ha in 1996 to 175 million ha in 2013 worldwide (Nap et al., 2003). It contributed substantially towards economic, health, longer shelf life, social and environmental benefits, particularly for poor farmers from developing countries and promotes the financial gain for alleviation of poverty. However, we have to be more careful regarding cross contamination, which may produce super weeds or may produce allergic substances in the growing environments. Moreover, not enough testing and research done on genetically modified foods and the long-term effects have not been discovered yet. This makes many people feel uneasy at random use of GM foods.Salinity seems to plays a key role in signaling to up regulate proline biosynthesis pathway and osmotin like protein has been identified from intercellular space of a haplotype- Mesembryanthemum crystallinum L. and association of osmotin protein with tonoplast of tobacco is known (Singh et al., 1989 and Yen et al., 1994). Genes encoding osmotins have numerous functions in improving stress tolerance (Jami et al., 2007), therefore, the over-expression of osmotin genes in transgenic rice increase the tolerance level against salinity and cold stresses or both in combinations (Goel et al., 2010). Though the mechanism of osmotin for mediating plant response to abiotic stress is still not so clear, primarily for salt tolerance it acts by reducing the build-up of Na+ ions in the cytoplasm or acting as Na+/H+ anti-porter has been recognized (Le et al., 2018). This evidence advocates that osmotin plausibly take part in intercellular compartmentalization of Na+ ions in vacuole and intercellular space to minimize deleterious effect of excess ion concentration in cytoplasm through reduced uptake of Na+ in cell and help restore membrane stability (Yen et al., 1994). Similarly, ROS like ascorbate peroxidase (APX) found to be accumulated in response to various abiotic stresses by creating an intrinsic antioxidant system and it plays a crucial role in elimination of H2O2. It is reported that when transgenic plants harbouring osmotin gene are challenged by abiotic stresses (like cold stress), APX gene expression increased substantially in the cytosol and ascorbate content increases but non transgenic plants were found to accumulate less ascorbate (Patade et al., 2013). Osmotin could be involved in osmotic adjustment of the cells by facilitating accumulation or compartmentalization of solutes (Barthhakur et al., 2010). It also protects the native structure of proteins during stress and for repairing of denatured proteins too. During our experimentation, the negative control plants when exposed to high salt concentration (NaCl), greenish leave sturned to yellow due to reduction of chlorophyll content. This reduction of chlorophyll content is associated with interaction of Na+ and Cl- ions with the enzymes involved in chlorophyll biosynthesis (Husaini & Abdin, 2008) [Data not presented]. Transgenic rice when exposed to high NaCl concentration, over expressed osmotin, restored high chlorophyll content through accumulation of proline that may reduce the ROS and induces ribulose oxygenase activity. Some reports showed that osmotin could enhance abiotic tolerance in plants by modulating stress responsive genes and by upregulating the levels (Patade et al., 2013), which was our main focus in indica rice in the present experiment. Osmotin could activate heteromeric G protein and mitogen-activated protein kinase (MAPK) proteins that responsible for catalyzing phosphotransfer reactions also play a role as a regulator in plant stress tolerance via cell signalling (Viktorova et al. 2012). These reactions are elemental to most signaling and regulatory pathway associated with enzyme activation, protein localization, macro-molecule assembly, and degradation as plant stress responses. Production of transgenic indica rice in our present study prospects to undertake rice husbandry constrained with high NaCl in the low-lying rice growing coastal areas that paves a way out to benefit Andaman Islands and elsewhere endowed with similar problems of saline soils. In nutshell, it may be concluded that this line should be tested along with a few other promising salt tolerant high yielding lines more rigorously. It is expected that, appreciable tolerance towards excess NaCl may be developed in Annada background in future for large–scale cultivation on saline soils in Andaman’s and elsewhere.
ACKNOWLEDGEMENTS
- The authors are thankful to Dr. K.C. Bansal, National Research Centre for Plant Biotechnology, Indian Agricultural Research Institute, New Delhi 110012 for providing the osmotin gene construct and to the Director, Central Agricultural Research Institute (presently CIARI), Port Blair for his keen interest, constant encouragement and facilities provided for conduct of this work.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML