-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Advances in Life Sciences
p-ISSN: 2163-1387 e-ISSN: 2163-1395
2016; 6(3): 49-53
doi:10.5923/j.als.20160603.01

Bacteriological Quality of Drinking Water Sources of Hail, Saudi Arabia
Abdullah Sulaiman Alshammari, Abdel Moneim Elhadi Sulieman, Vajid Netoor Veettil, Abdelmuhsin Abdelgadir Abdelmuhsin
Department of Biology, Faculty of Science, University of Hail, Kingdom of Saudi Arabia
Correspondence to: Abdel Moneim Elhadi Sulieman, Department of Biology, Faculty of Science, University of Hail, Kingdom of Saudi Arabia.
| Email: |  |
Copyright © 2016 Scientific & Academic Publishing. All Rights Reserved.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

This study aimed to detect the presence of pathogenic bacteria in drinking water sources of Hail, Saudi Arabia and to characterize and identified these bacteria. A total of 90 water samples were collected from 10 different sites of Hail area. To find out pathogenic bacteria, culturing technique was used followed by staining for identification of bacterial isolates. Then Phoenix system was used for identification of bacterial species. Some of the tested samples contained pathogenic bacteria, especially those of natural. Five different bacterial species were found including Escherichiacoli (E.coli), Psuedomonasaeruginosa(P.aeruginosa), Salmonella, Klebsiella, Staplylococcus and Enterobacteraerogenes. E.coli was mostly isolated species that was identified in 24 samples (26.66%) followed by P.aeruginosa: 11 samples (12.22%), Klebsiella: 8 samples (8.88%) and Salmonella: 6 samples (6.66%). This study concludes that disinfection of water should be implemented to reduce water borne diseases, water supplying departments have to follow WHO standards for better public health and to control disease outbreak by coliforms.
Keywords: Contamination, Drug resistance, E.coli, P.aeruginosa, Salmonella, Klebsiella
Cite this paper: Abdullah Sulaiman Alshammari, Abdel Moneim Elhadi Sulieman, Vajid Netoor Veettil, Abdelmuhsin Abdelgadir Abdelmuhsin, Bacteriological Quality of Drinking Water Sources of Hail, Saudi Arabia, Advances in Life Sciences, Vol. 6 No. 3, 2016, pp. 49-53. doi: 10.5923/j.als.20160603.01.
Article Outline
1. Introduction
- Water is considered a vehicle for the propagation and dissemination of human associated bacteria. Water is an important chemical molecule containing feature of life it can be dissolved into organic compounds, salts, inorganic compounds and gases that are involved in metabolic processes because it is universal solvent and due to that it provides stability to membrane system, macro molecules, transportation and thermal regulation of body [1-4]. The world Health Organization [5] defined safe drinking water as that water having acceptable quality in terms of its physical, chemical and bacteriological parameters. Bacteriological parameters especially Escherichia coli (E.coli) and total coliform have been used to determine the general quality of drinking water in the world [5, 6]. Pathogenic contamination of water is also important threat for living organisms. In Asian regions peoples those are living near to rivers are at high risk of their lives because of sewage pollution which is directly disposed from chemical factories and septic tanks that are the main reservoir of pathogens involves in water borne diseases [7]. Developing regions lack in provision of safe drinking water to their peoples and in Africa and Asia almost 800 billion individuals using unsafe drinking water which results in suffering of individual from water borne diseases [8].In Saudi Arabia, the quality of drinking water is currently receiving some attention from environmentalist and water scientists. Although, most regions in Saudi Arabia do not face serious problems regarding drinking water as other parts of world, there are still water quality problems, particularly the microbiological [9-11].Waterborne disease is a global burden which is estimated to cause more than 2.2 million deaths pear year and higher cases of illness every day, including diarrhea, gastrointestinal diseases and systematic illnesses [12, 13]. Worldwide, an economic loss of nearly 12 billion US dollars per year is estimated [14]. Waterborne infections are caused by ingestion, airborne or contact with contaminated water by a variety of infectious agents which includes bacteria, viruses, protozoa and helminths [15]. Although microorganisms in drinking water are reduced by chlorination, they may survive the treatment process and enter the distribution system. Moreover, the presence of antibiotic resistance in microorganisms has been previously reported [16-18]. The present study was designed to investigate the prevalence of bacterial pathogens, to characterize and to identify these bacteria.
2. Materials and Methods
2.1. The Study Area
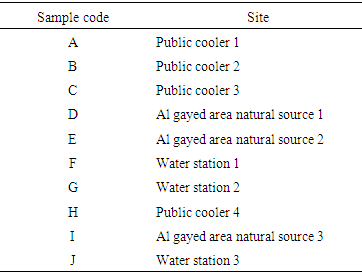
- The study was conducted in some sites of Hail area. Hail city is located in the middle of the northern region of the Arabian Peninsula, between 25°-29° N and 38°-42° and has an average elevation of 900-1350m above sea level. It is one of the major cities in the Kingdom of Saudi Arabia and considered as the fifth city in regard to its area. The local geology is dominated by the Arabian shield rock extending to steep wadies and hills. It is characterized by its limestone sand, which exists in the form of sand sheets and sand dunes.
|
2.2. Microbiological Analysis
2.2.1. Colony Count
- Total viable count was carried out using the pour plate technique as described by Harrigan and MacCance [19]. Ten ml of each sample were transferred to 90 ml of sterile Ringer’s solution, as a first dilution 101, serial dilutions were made up to 106 and 1ml of each dilution was transferred aseptically in duplicate into sterile Petri-dishes. 10- 15 ml of melted plate count agar were poured into the dishes. The dishes were then thoroughly mixed to facilitate distribution of the sample throughout the medium, the medium was allowed to solidify and plates were incubated at 370°C for 48 hours. Colony counter (Labtech) and hand-tally were used for the determination of the total bacterial counts in terms of colony forming units per ml (c.f.u./ml).
2.2.2. Isolation by Membrane Filtration
- For all samples, three volumes of 100 mL were filtered through 0.45 µm pore sized filter (cellulose nitrate membranes, Millipore corporation) using a water pump (model Rocker 4000). These membranes were aseptically placed up on plates with appropriate selective media ensuring that no air bubbles were trapped. The selective media used are as follows. mFC agar used as a selective medium for faecal coliforms, mEndo for total coliforms. All the media were prepared according to the manufacturers’ instructions (Himedia, Merck). Each sample was analysed in triplicate. The colonies were enumerated, characterized, and recorded. The results were expressed as the number of faecal coliforms, total coliforms in 100 mL of water. Blue colonies from mFC agar (presumptive coliforms), metallic-sheen colonies from mEndo agar (presumptive total coliforms).
2.2.3. Purification of Colonies
- Colonies were purified by twice sub-culturing using the streaking plate method. Young cultures were used for Gram staining and all isolates were identified as Gram-negative Bacilli. All the Gram-negative isolates were subjected to primary and secondary biochemical identification tests.
2.2.4. Oxidase Test
- Oxidase regent was prepared using manufacturer instructions, and then 2-3 drops of it were poured on filter paper placed in petri plate. The bacteria which changed the color to deep purple on treated filter paper within 10 seconds were report as oxidase +ive.
2.2.5. Detection of Extended Spectrum B-Lactamase (ESBL) Production
- Detection of ESBL phenotype was carried out using BD phonix ESBL automated system (Becton, Dickinson, Md., USA). A total of 20 isolates were used in this test. The isolates were sub-cultured on Mac-Conkey agar to obtain pure culture for which a 0.5 McFarland suspension was obtained and tested according to the manufacturer provided protocol.
2.2.6. Phoenix Analysis
- The isolated bacteria were further identified by the BD Phoenix Automated Microbiology System (PAMS, MSBD Biosciences, Sparks, MD, USA), employing the Phoenix TM NMIC/ID Panels according to the manufacturer’s instructions. Susceptibility of the isolated bacteria to antimicrobial agents was determined by the disc-diffusion methods according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) [20]. Susceptibility of staphylococci to methicillin was determined by the cefoxitin disc-diffusion method. The following quality control organisms E. coli ATCC 25922, K. pneumoniae ATCC 700603, and Pseudomonas aeruginosa ATCC 27853 were used with gram-negative bacilli and S. aureus ATCC 29213 and S. aureus ATCC 25923 with staphylococci.
3. Results and Discussion
3.1. Microbiological Characteristics of Water Samples
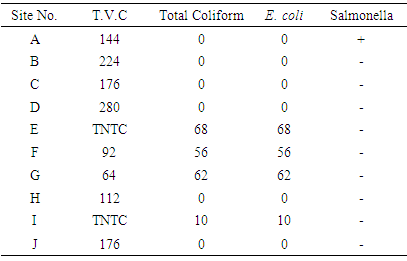
- The results of the microbiological analysis of water samples collected from various sites are presented in Table (2). The results indicate that 4 sites (40%) of tested water samples show the presence of total coliform, and Eschrichia coli (E. coli) were detected in these samples indicating unsuitability of water samples in these sites for drinking and/or food preparation according to the International Standards [21] for drinking water, which stated that E.coli or thermotolerant coliform bacteria and pathogenic intestinal protozoa must not be detectable in any 100 ml sample. Generally Coliform bacteria originate in feces (e.g. Escherichia) as well as genera not of fecal origin (e.g. Enterobacter, Klebsiella, Citrobacter). These bacteria are intended to be an indicator of faecal contamination; more specifically of E. coli which is an indicator microorganism for other pathogens that may be present in faeces. Presence of faecal coliforms in water may not be directly harmful, and does not necessarily indicate the presence of faeces [21]. Water quality is monitored in many countries such as U.S. and Canada to protect the health of the general public. Bacteria contamination is one monitored pollutant. In the U.S., faecal coliform testing is one of the nine tests of water quality that form the overall water-quality rating in a process used by U.S. The faecal coliform assay should only be used to assess the presence of faecal matter in situations where faecal coliforms of non-faecal origin are not commonly encountered (Doyle and Erickson [22]. EPA [23] has approved a number of different methods to analyse samples for bacteria.
|
3.2. Identification of Bacteria
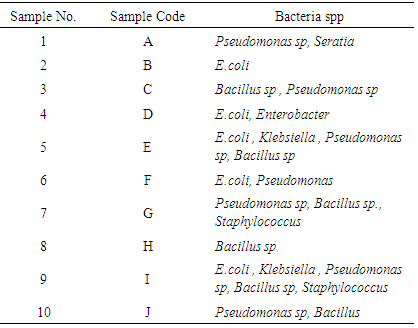
- Despite the advent of the rapid microbial identification tools – particularly those based on mass spectrometry and nucleic acid-based identification techniques since they are widely used for detecting and identifying microbes in clinical and environmental samples.The bacterial species dominated the various water samples are shown in Table (3). The bacterial species were tentatively identified as Pseudomonas sp, Seratia, E.coli, Enterobacter, Klebsiella and Bacillus sp.
|
4. Conclusions
- In conclusion, water which has not been subjected to treatment in some sites of Hail area was contaminated with various bacterial species, so it is not sutable for drinking or preparation of food. The findings of the present study demonstrate the need to come up with source water protection strategies and policies for rural communities in some of which, water treatment is not available. Public awareness about the importance of protecting water resources as well as monitoring their quality and effects on human health are also important, and recommended.
ACKNOWLEDGEMENTS
- The authors would like to thank the University of Hail for the financial support to carry out this project (SCB-114).
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML