-
Paper Information
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Advances in Life Sciences
p-ISSN: 2163-1387 e-ISSN: 2163-1395
2016; 6(1): 1-6
doi:10.5923/j.als.20160601.01

Glycoside Hydrolase Family 43 from Paenibacillus curdlanolyticus Strain B-6; An Accessory Enzyme to Enhance the Hydrolysis of Pretreated Rice Straw
Kanok Wongratpanya1, Junjarus Sermsathanaswadi2, Thidarat Nimchua3, Rattiya Waeonukul4, Patthra Pason4, Chakrit Tachaapaikoon4, Akihiko Kosugi5, Khanok Ratanakhanokchai1
1School of Bioresources and Technology, King Mongkut’s University of Technology Thonburi (Bangkuntien Campus), Bangkok, Thailand
2Department of Chemical Technology, Faculty of Science and Technology, Suan Dusit Rajabhat University, Bangkok, Thailand
3Enzyme Technology Laboratory, Bioresources Technology Unit, National Center for Genetic Engineering and Biotechnology (BIOTEC), Thailand Science Park, Pathumthani, Thailand
4Pilot Plant Development and Training Institute, King Mongkut’s University of Technology Thonburi (Bangkuntien Campus), Bangkok, Thailand
5Biological Resources and Post-harvest Division, Japan International Research Center for Agricultural Sciences (JIRCAS), Ibaraki, Japan
Correspondence to: Khanok Ratanakhanokchai, School of Bioresources and Technology, King Mongkut’s University of Technology Thonburi (Bangkuntien Campus), Bangkok, Thailand.
| Email: |  |
Copyright © 2016 Scientific & Academic Publishing. All Rights Reserved.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/

Glycoside hydrolase family 43 (GH43) from Paenibacilluscurdlanolyticusstrain B-6 (Xyl43B6) was preformed heterologous expression in Escherichiacoli. Enzymatic function determination revealed that Xyl43B6 exhibited trifunctional properties of α-L-arabinofuranosidase, β-xylosidase, and endo-xylanase. Xyl43B6 showed broad specificity of substrates on aryl-arabinoside, aryl-xylopyranoside, xylooligosaccharides, xylobiose, low branching xylan, and highly substituted arabinoxylan. Xylose liberation rate from xylooligosaccharides was higher than from xylobiose. However, trace amount of xylose was detected from highly substistuted rye flour arabinoxylan, indicating influence of chain length and substitution groups on xylose liberation efficiency of Xyl43B6. Moreover, co-hydrolysis of Xyl43B6 with GH10 endo-xylanase (Xyn10E) on pretreated rice straw revealed the boosting effect and increase in liberated reducing sugar as a result of Xyl43B6/endo-xylanase synergy. This report proposed new enzymatic properties of GH43 which showed the ability to enhance biomass biorefinery efficiency.
Keywords: α-L-Arabinofuranosidase/β-xylosidase/xylanase activities, Glycoside hydrolase family 43, Multifunctional enzyme,Paenibacilluscurdlanolyticus, Rice straw biorefinery
Cite this paper: Kanok Wongratpanya, Junjarus Sermsathanaswadi, Thidarat Nimchua, Rattiya Waeonukul, Patthra Pason, Chakrit Tachaapaikoon, Akihiko Kosugi, Khanok Ratanakhanokchai, Glycoside Hydrolase Family 43 from Paenibacillus curdlanolyticus Strain B-6; An Accessory Enzyme to Enhance the Hydrolysis of Pretreated Rice Straw, Advances in Life Sciences, Vol. 6 No. 1, 2016, pp. 1-6. doi: 10.5923/j.als.20160601.01.
Article Outline
1. Introduction
- Lignocellulosic biomass is the most abundant renewable resource obtained from agricultural residues, herbaceous grasses and forest products. Generally, lignocellulosic biomass is composed of approximately 15-40% cellulose, 10-30% hemicelluloses and pectin, and 5-20% lignin. [1] Xylan is the major component of hemicelluloses encompassing a linear β-(1,4)-D-xylopyranose backbone decorated with different groups of side chain. Common substituents found on the backbone of xylan are acetyl, arabinosyl, and glucuronosyl residues. Furthermore, type and degree of the substituted side chains vary among botanical origins. [2] Due to the recalcitrant architecture of xylans, hydrolysis of xylans requires the action of a multiple-enzyme system. The group of xylanolytic enzymes works synergistically to depolymerize xylan to its sugar constituents. Xylanolytic enzymes are usually composed of non-debranching enzymes (endo-xylanase and β-xylosidase) and debranching enzymes (α-arabinofuranosidase, α-glucuronidase, acetyl xylan esterase and phenolic acid esterase). [2]A concept of xylan bioconversion has been emphasized in the last few decades. Thus, effective utilization of xylan would play a significant role in economical production of highly valued substances. Xylan bioconversion required a combination of debranching and non-debranching enzymes. Releasing the substituted side groups from xylan allows endo-xylanase to act efficiently for complete hydrolysis. [2] The glycoside hydrolase family 43 plays a role as an accessory enzyme that boosts the hydrolysis efficiency of endo-xylanases. GH43 is a group of enzymes, encompassing monofunctional enzymes such as α-L-arabinofuranosidase (EC 3.2.1.55), β-xylosidase (EC 3.2.1.37), arabinanase (EC 3.2.1.99), endo-xylanase (EC 3.2.1.8), galactan 1,3-β- galactosidase (EC 3.2.1.145), α-1,2-L-arabinofuranosidase (EC 3.2.1.-), exo-α-1,5-L-arabinofuranosidase (EC 3.2.1.-), exo-α-1,5-L-arabinanase (EC 3.2.1.-), and β-1,3-xylosidase (EC 3.2.1-). Furthermore, multifunctional enzymes, such as β-xylosidase/α-L-arabinofuranosidase [3], β-xylosidase/ exo-xylanase (GenBank ID: ABD48561.1), and β-1,4- xylosidase/α-1,5-arabinofur(pyr)anosidase/β-1,4-lactase/α-1,6-raffinase/α-1,6-stachyase/β-galactosidase/ α-1,4-glucosi-dase [4], have been reported.Paenibacillus curdlanolyticus strain B-6 is a cellulolytic/ xylanolytic bacterium that produces a unique extracellular xylanolytic-cellulolytic multienzyme complex-like structure capable of degrading insoluble substrates. [5] We found one open reading frame (ORF) from the B-6 strain encoding a protein belonging to GH family 43 which comprises a broad variety of enzyme specificities and similarities. [6] We speculated that this GH43 may be a good accessory enzyme in xylanolytic enzymes cocktail that boosts the hydrolysis efficiency for the bioconversion of biomass. Therefore, synergy analysis was elucidated in this study.
2. Materials and Methods
2.1. Strains and Plasmid
- P. curdlanolyticus B-6 was isolated from an anaerobic digester and fed with pineapple waste. [5] All cloning strategies were performed in Escherichia coli DH5α (New England Biolabs, Ipswich, MA, USA). E. coli BL21 (DE3) (Novagen, Darmstadt, Germany) was used as the host for the derivative of pET28b(+) expression system (Novagen).
2.2. Gene Cloning and Expression
- The pXyl43B6 plasmid construction and Xyl43B6 expression were conducted as described by Wongratpanya et al. [6]. Protein purification by affinity chromatography method was achieved via HisTrapTM FF columns (GE healthcare, Little Chalfont, UK). Protein concentration was determined as described by Lowry et al. [7].
2.3. Enzyme Assays
- α-L-Arabinofuranosidase and β-xylosidase activities were assayed on p-nitrophenyl-α-L-arabinofuranoside (pNPA) and p-nitrophenyl-β-D-xylopyranoside (pNPX), respectively (Sigma- Aldrich, St. Louis, MO, USA), as described previously. [6] One unit (U) of enzyme activity was defined as the amount of enzyme required for liberating 1 µmol of p-nitrophenol per min. Xylanase activity was assayed on 1% xylan from birchwood (Sigma-Aldrich) in 50 mM sodium phosphate buffer (SPB) (pH 7.0) at 50°C for 10 min. Liberated reducing sugars were analysed by the Nelson-Somogyi method [8]. One unit (U) of xylanase activity was defined as the amount of enzyme used for liberating 1 µmol of xylose per minute.
2.4. Substrate Specificity
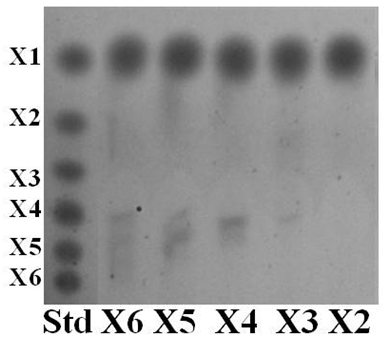
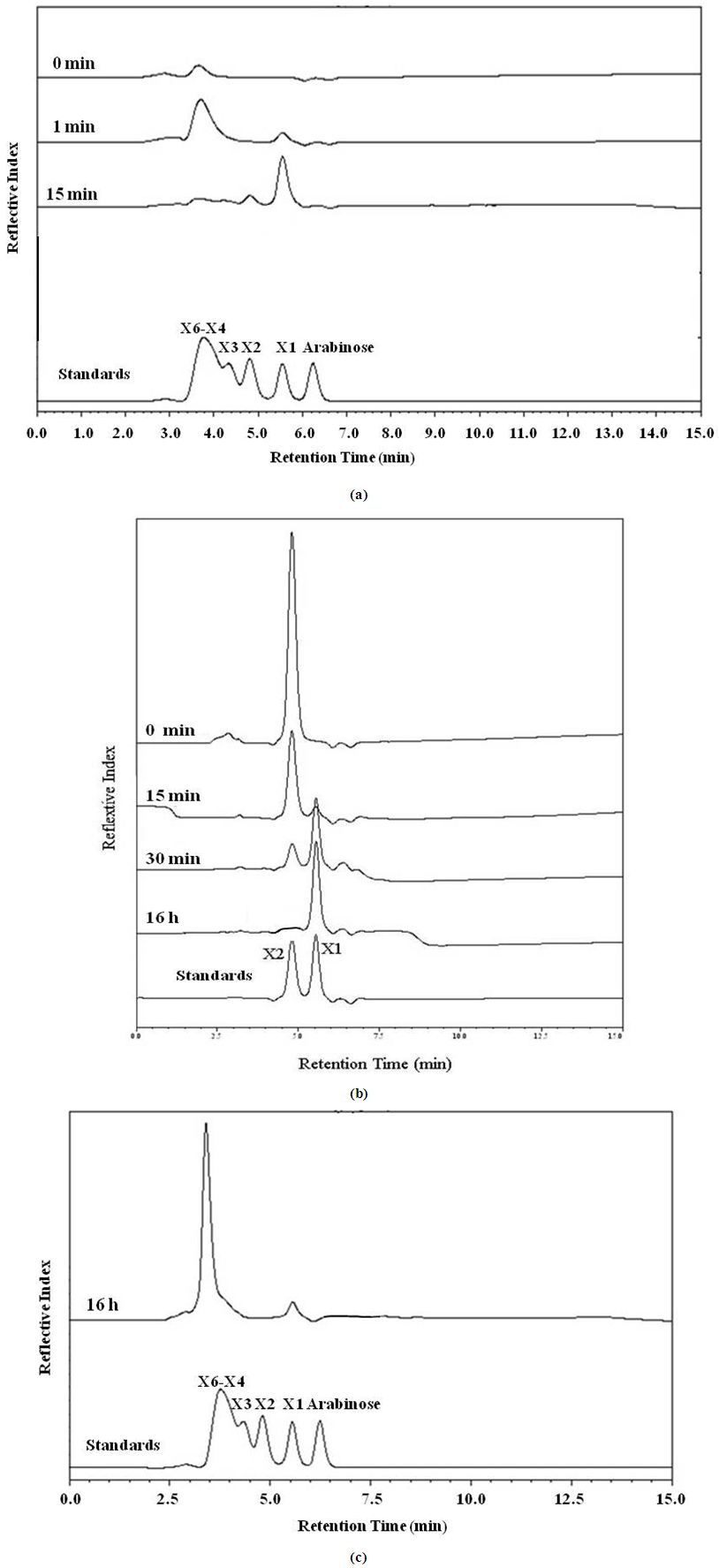
- Hydrolysis of Xyl43B6 was determined on various structural types of substrates which were: 0.025% (w/v) xyloligosaccharides (X3-X6) and xylobiose (Megazyme International, Wicklow, Ireland), 1% (w/v) xylans from birchwood and oat spelt (Sigma-Aldrich), wheat flour arabinoxylan and rye flour arabinoxylan (Megazyme). The hydrolysis products were separated by thin layer chromatography (TLC), as described by Sornyotha et al. [9] and high performance liquid chromatography (HPLC) (Shimadzu, Kyoto, Japan) with a reflective index detector (Shimadzu RID-10A) on BP-100 Pb++ carbohydrate columns (Benson Polymeric, Sparks, NV, USA) operated at 85°C with deionised water as a mobile phase at a flow rate of 0.6 mL/min.
2.5. Biomass Hydrolysis
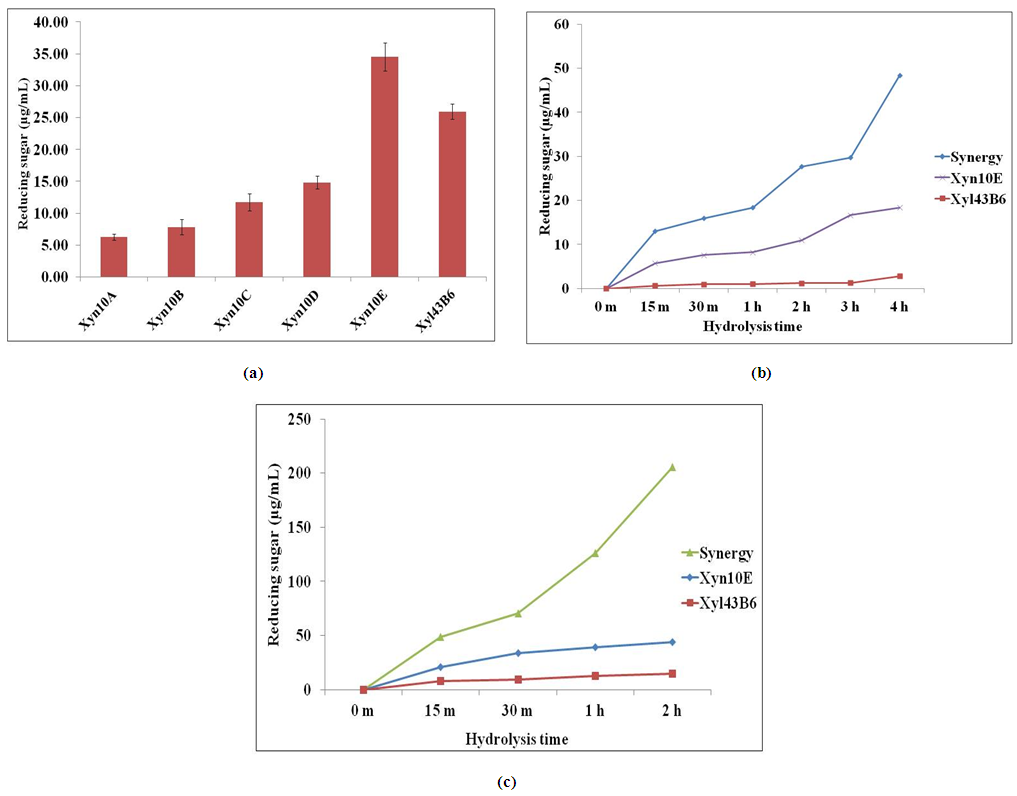
- Enzymatic hydrolysis of biomass was performed using 1% grinded rice straw at pH 7.0 using 50 mM SPB at 50°C. One nmol of enzymes (each): Xyl43B6, Xyn10A [10], Xyn10B [11], Xyn10C [12], Xyn10D [13], and Xyn10E (unpublished data) was mixed and incubated for 16 h. The liberated reducing sugars were quantified by the Nelson-Somogyi method.
2.6. Synergistic Analysis on Rice Straw Hydrolysis
- Pretreatment of rice straw was carried out by soaking in 27% ammonium hydroxide at a solid:liquid ratio of 1:10 at 60°C for one week. The sample was neutralized by 1 N HCl and the pretreated solid was retrieved, dried, milled by a blender and used as a substrate for enzymatic hydrolysis. The synergistic effect of Xyl43B6 and GH10 endo-xylanase was studied by using 1% (w/v) of pretreated rice straw in 50 mM SPB (pH 7.0). One nmol of each enzyme was mixed with the substrate and incubated at 50°C. Liberated reducing sugars were quantified by the Nelson-Somogyi method.
3. Results and Discussion
- According to Fosmid genomic library of strain B-6 constructed previously [11], an ORF Xyl43B6 encoding a protein was confidently predicted by SMART software [14] to belong to a glycoside hydrolase family 43 catalytic domain. Heterologous expression of Xyl43B6 was accomplished in E. coli. Protein expression and affinity purification was achieved via either C and N terminals histidyl tag, originated by BamHI/XhoI insertion site on pET28b(+) expression vector. A single protein band of approximately 39 kDa was observed on SDS-PAGE analysis.Purified Xyl43B6 was determined the enzymatic hydrolysis function in various kinds of substrates. Xyl43B6 showed hydrolysis capability on pNPA and pNPX with the same activity (0.30 U/mg). Furthermore, determination of xylanase activity showed that Xyl43B6 exhibited xylanase activity versus various structural types of xylan. The highest xylanase activity (2.86 U/mg) was achieved on birchwood xylan, and 1.46 U/mg was determined on oat spelt xylan. Moreover, weak xylanase activity of Xyl43B6 on recalcitrant highly substituted xylan, wheat flour arabinoxylan and rye flour arabinoxylan was detected. As showed in Fig. 1, xylose was found as a sole hydrolysis product from xylooligosaccharides X3-X6 and xylobiose. Those results confirmed that Xyl43B6 exhibited multiple functions of hydrolysis activity including α-L-arabinofuranosidase, β-xylosidase and xylanase with broad substrate preference. Xyl43B6 could hydrolyze xylobiose, xylooligosaccharides, low branching xylan, and highly substited xylan. However, hydrolysis efficiency on highly substituted xylan was low. It might be postulated that increasing structural complexity of xylan had an effect on hydrolysis efficiency of Xyl43B6. On the other hand, β-xylosidse function of Xyl43B6 was different from true β-xylosidases which prefer xylobiose only [15]; the hydrolysis efficiency decreases with increasing degrees of polymerisation. [16]
4. Conclusions
- In summary, an ORF Xyl43B6 from the strain B-6 genomic fosmid library was heterologously expressed in E. coli. Xyl43B6 exhibited multifunctional enzymatic activities with broad substrate specificity. Xyl43B6 plays a role as an accessory enzyme that boosts liberation of reducing sugar of GH10 endo-xylanase on rice straw hydrolysis. Hence, co-hydrolysis of Xyl43B6 with endo-xylanase could improve biomass biorefinery efficiency.
ACKNOWLEDGMENTS
- This work was financially supported in part by King Mongkut’s University of Technology Thonburi, Thailand (under the Higher Education Research Promotion and National Research University Project of Thailand, Office of the Higher Education Commission), and Thailand Graduate Institute of Science and Technology.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML