-
Paper Information
- Previous Paper
- Paper Submission
-
Journal Information
- About This Journal
- Editorial Board
- Current Issue
- Archive
- Author Guidelines
- Contact Us
Advances in Life Sciences
p-ISSN: 2163-1387 e-ISSN: 2163-1395
2014; 4(3): 123-130
doi:10.5923/j.als.20140403.05
Identifying Probiotics of Macrobrachium rosenbergii’s Intestine Micro Flora and Their Effect in Prawn’s Growth and Survival
Bahareh Rahimian Zarif1, Mehrdad Azin2
1Department of Biology, Sanandaj Branch, Islamic Azad University, Sanandaj, Iran
2Biotechnology Center, Iranian Research Organization for Science and Technology, P.O. Box 15815-3538, Tehran, 15819, Iran
Correspondence to: Bahareh Rahimian Zarif, Department of Biology, Sanandaj Branch, Islamic Azad University, Sanandaj, Iran.
| Email: |  |
Copyright © 2014 Scientific & Academic Publishing. All Rights Reserved.
Introduction: In this research, probiotics of freshwater prawn (Macrobrachium rosenbergii)’s intestine micro flora that affected to growth and survival were isolated.Method: after doing identifying tests, isolation of micro flora bacteria was done and with examining probiotic properties, 4 bacteria were recognized that with different quantities separately or mixture together was added to prawns diet. After 60 days, effects of probiotics on biometric parameters were recognized. Effective bacteria in this variation, identified by gene sequencing with 16srRNA and drawn phylogenic tree. Results: adding of equal quantities of S202 and S203 synchronously, in pellet form to prawns diet, was increased body weight percent, total length percent, Carapace length percent, Daily Growth Average percent and Specific Growth Rate percent significantly (p<0.05). After gene sequencing with 16srRNA and drawing phylogenic tree of S202 and S203, defined these bacteria belong to Bacillus lichniformis and fecium Entrococcus strains. Discussion and conclusion:whereas Macrobrachium rosenbergiiis important commercially due to having pleasant flavor meat, the least level of cholesterol and high level of protein, using the best and low cost way for high production of this prawn is important that access to this object is possible with using probiotics in prawn diet.
Keywords: Probiotic, Macrobrachium rosenbergii, 16srRNA، Bacillus lichniformis, Entrococcus fecium, Prawn
Cite this paper: Bahareh Rahimian Zarif, Mehrdad Azin, Identifying Probiotics of Macrobrachium rosenbergii’s Intestine Micro Flora and Their Effect in Prawn’s Growth and Survival, Advances in Life Sciences, Vol. 4 No. 3, 2014, pp. 123-130. doi: 10.5923/j.als.20140403.05.
Article Outline
1. Introduction
- Macrobrachium rosenbergii or huge size prawn in the river, is one of the first species that have been scientifically identified.Rosenbergii species is a sample of turbid water prawns and its geographical distribution is more within the sweet waters where its temperature average during the day and night is partially fixed. This species is the most important farmed prawn of sweet water which for Commercial production was transferred to the most countries of the world.Commercial importance of this prawn is for the favorite taste of meat, low cholesterol level and also due to the high protein storage level. [1]According to the use of approaches which can increase the generation of this aquatic creature in a large scale is very important and in this context adding the live bacteria as probiotics to the food of prawn is very successful.These bacteria are resistant to the Acidic pH and intestinal bilious solutions and due to this reason are located in this region and having the advantages like the vitamins production and other requisite nutrition, digestion of complex nutritional materials, reinforcing the immune system and mucous secretion. [1]These bacteria are very various regarding their genus and species and often located in the species of Bacillus, Lactococcus, Micrococcus, Enterococcus, lacobacillus, Streptcoccus Aeromonas, Photorhodo bacterium, Pseudomonas, and Vibrio.The identification of species and associated species can be performed by 16S rRNA gene sequencing. In this research also genus and species of probiotics effective upon the biometric indices via this technique was identified.
2. Methods and Materials
2.1. Sampling and Bacterial Isolation
- Bacterial isolates in sterile conditions after transfer of prawns (M. rosenbergii) from freshwater prawn ponds located in the city of Qasr-e Shirin in Kermanshah province to the research laboratory of Islamic Azad University of Sanandaj taken from the intestine of 100 prawns. The intestinal samples were cultured on liquid medium as (Brain Heart Infusion) BHI and incubation was performed at 37°C for 48-24 hours and the bacterial purification after dilution on nutrient agar media such as ships, MRS agar (DeMan, Rogosa Sharpe Agar), M17 and Seawater Agar (SWA) was done and after incubation at 37-35°C for 48-24 hours, the initial identification of the purified bacteria using Gram stain was performed under the sterile conditions and biochemical tests (2,4-5).
2.2. Characterization of Probiotic Bacteria
2.2.1. Resistance to Acid and Bile Salts
- Evaluation of bacteria's tolerance isolated from intestinal towards acid and bile salts was performed through providing conditions that was relatively similar to the environmental conditions of prawn's intestine. 1 ml of a 48 -24 hour culture of bacteria isolated in isotonic buffer (Bott and Wilson salts) containing (grams per hundred milliliters): 1.24 percent of potassium hydrogen phosphate, 0.76% of Potassium dihydrogen phosphate, 0.1% of Tri-Sodium citrate, 0.6% of ammonium sulfate with 50% of the sodium Chelate and sodium Chelate di-oxy with 6.7 pH, incubated with agitation at 37°C. Bile resistance by culturing samples were analyzed after 3 hours. To study the characteristics of resistance to acid, bacteria in isotonic buffer containing 85% w - v of sodium chloride and 1 mg/ml of pepsin with 2 pH implemented in suspension and incubated with stirring at 37°C.Resistance to the acid in this type of bacteria was investigated during 30min-3h. The various dilutions of bacteria resistant to the acid and bile directly upon the nutrient gar culture and MRS agar was cultivated, and incubated during 24 h at 37°C and CFU of bacteria as measured [6].
2.2.2. Antimicrobial Properties
- One of the properties that probiotics should be having, is the antimicrobial properties against the certain gram-positive and gram-negative bacteria.The antibacterial impacts of bacterial species separated from prawn's intestine via the agar diffusion method was measured upon the indicator bacteria of Staphylococcus، Aeromonas MRCS 2 and Vibrio alginolyticus MCCB 112. [7].Probiotic bacteria separated from were cultured within the environments of MRS broth and BHI broth at 37°C and 24h,Then each of bacterial culture environments at 5340× g for 15 min at 4°C were centrifuged and upper liquid was separated and upper liquid was used in 3 forms.A. upper fluid without upper material (CS)B. the upper clear fluid that its pH reached to 6.5-7 by addition of 1mol. (CSB)C. the upper transparent that its pH reached to 6.5-7 by addition of 1mol and also 200unit/ml catalase was added to it. (CSBC)The upper fluid without of added materials and also along with added materials within the sterile conditions was filtered by the membranes with 0.22 micrometer pores and these filters directly as used for study of antagonistic effects. The plates was filled with 20 ml by MHA and each of indicator bacteria was cultured with final concentration of 106 separately upon this environment.5 wells with diameter of 5.8 mm were created on the culture environment and 3 of them were filled with 85 micro-liters of CS fluid, CSB and CSBC which was mentioned it above and two other wells were considered as the positive and negative controls.The negative controls, with sterilized MRS broth and positive control wells were filled with of chloramphenicol antibiotic. The inseminated plates were incubated at a temperature of 25℃ for 24 hours. The diameter of region without bacterial growth as measured by Caliper. [6-9]
2.2.3. Antibiotic Sensitivity
- The resistance or separated probiotic sensitivity compared to cephalothin, co-trimoxathin, cephalexin, lincomycin, amoxicillin, ampicillin, Cloxacillin, Novobiocin, penicillin G, tetracycline, chloramphenicol, gentamicin, Ciprofloxacin, erythromycin, Rifampicin, kanamycin,Vancomycin (), was investigated via agar disc diffusion method upon the culture of Oxoid Muller-Hinton(MH) [9].In this method, the bacterial suspension containing 108 CFU/mL bacteria that corresponded with McFarland tube-5, was spread on the Mueller Hinton agar culture medium using the Sterile swabs and antibiotic discs was located periodically upon it and after 16-24 h, growth lack 's halo diameter as investigated [6].
2.2.4. Measurement of the Binding to Cell Rate
- To investigate this characteristics of separated probiotics, was used of one-layer Caco-2 cells. First, these cells were washed twice with PBS. Then 1 ml of the bacterial suspension that containing 108 bacteria was added to the Caco-2 medium, then 2 ml of this suspension as added to each chamber of to-chamber slide.And at 37℃ for 60 min was incubated and after 1h incubation, mono-layers 2 times were washed with sterilized PBS and 1.2 pH and after the cells were fixed with methanol and stained with warm color, the bacteria connected to the cell were counted randomly at 60 microscopic points under the microscope and with lens-100 [9].
2.3. Test Conditions for Prawn
- 200 prawns with weight of 18.43 g and average body length of 11.58 cm were transferred to the prawn ponds in Gasre Shirin (Kermanshah) to the Glass aquariums in dimensions of (60 ×20 × 40 cm) in Research Laboratory of Sanandaj University.Each aquarium equipped to the heating system environmental pH was measured with pH meter daily which was about 22±2. 10 prawns were maintained in each aquarium. Biometric parameters such as body length, carapace length, body weight was measured in the prawn before adding probiotics [1].
2.4. Prawn Diet
- Basic diets for prawns including the plant foods (carrots, lettuce) and meaty (chicken meat) as the minced meat and the pellet dried prawn,3-4 times a day for every 2% by prawn weight (a day, a meat and vegetable dish) was added to the aquarium every other day [1].
2.5. Adding the Probiotics to the Food of Prawns
- After the confirmation of probiotic characteristics of separated bacteria, the 108CFU/mL density was cultivated from each bacteria into the media of broth MRS and broth BHI.After incubation at 35-32℃ for 48-24 h, each culture media was centrifuged for 15 min × g 5500 rpm.Each of the bacterial cell deposition separately or combined together (in 50:50) to 10 g per 1 kg of feed was mixed with the feed. Probiotic foods 4-3 times a day, for 2% of weight of prawns was added to the aquarium [1].
2.6. Effect of Probiotics on Growth Parameters of a Prawn
- During the period of 60 days nurture of prawns, biometric parameters such as body weight, body length was measured in treated and control samples.- Specific growth rate Percentage (Specific Growth Rate %): it was calculated for each prawn based on the percentage increase in weight per day.
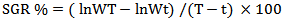 SGR%: the increase percentage of body weight per prawn per dayLn WT: natural logarithm of weight in a certain time (T)ln Wt: natural logarithm of initial weightT: given time:t: Initial time- (Percent Body Weight Increase %)
SGR%: the increase percentage of body weight per prawn per dayLn WT: natural logarithm of weight in a certain time (T)ln Wt: natural logarithm of initial weightT: given time:t: Initial time- (Percent Body Weight Increase %)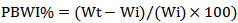 - Average Daily Growth %This value is calculated according to the weight of prawn per day.
- Average Daily Growth %This value is calculated according to the weight of prawn per day.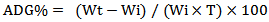 Wi: the initial weightWt: total weightT: length of growing period [10]
Wi: the initial weightWt: total weightT: length of growing period [10]2.7. Bacterial Identification through 16s rRNA
- Genomic DNA of Probiotics affecting the biometric parameters and the lifespan of the prawn using phenol / chloroform was extracted from a 24 hour culture the bacteria in LB broth medium [11].The 16S rRNA gene was amplified using primers fD1 (5'-agagtttgatcctggctcag-3') and rD1(5'-aaggaggtgatccagccgca -3') (12), which was designed by SinaClon.DNA proliferation using 50 microliters of compounds containing 5 microliter of 10 x PCR buffer, 2 microliter of a mixture of 10 mM of dNTP, 1.5 ml of the 50 mM MgCl2 solution, 2 micro liters (10 pico moles per micro liter) of each primer, 1 microliter (6-10)ng of DNA and 0.6microliter of the Taq DNA polymerase enzyme.PCR reactions through thermal cycler machine (Techne Flexigene, Model FFGO5TUD, Minneapolis, MN, USA) and with a temperature program of 92℃ for 1 min, 30 cycles of 95℃ for 1 min and 50℃ for 30 seconds, 72℃ for 1 min and 30 sec and 72℃ for 10 min was performed.The PCR products transferred toa 1% agarose gel, and then stained with ethidium chloride, and then imaging as done using the Gel document device.Primers pair, 27F (5'-agagtttgatcctggctca -3 ') and 1492R (5'-taccttgttacgacttcacc -3') which were complementary to the inner regions of the genome, was used for complete amplification of 16S rRNA gene and in this reactions, was used of the concentration of 2 mM MgCl2 (13) and for desired DNA fragments proliferation by these primers, PCR with a temperature schedule of 95℃ for 5 min followed by 30 cycles of 95℃ for 40 seconds for better denaturation of DNA molecule and temperature 57.6℃ for 90 s and a temperature of 72℃ for 90 s and a temperature of 72℃ for 5 min was used.DNA gene sequencing was done by Microgen Company in Korea. Review and analysis of gene homology by BLAST (Blastn algorithm) on the site http:// www.ncbi.nih.gov / took place.
3. Results
3.1. Early Identification of the Isolated Bacteria
- Early identification of bacterial species were performed using the differential media and biochemical tests under the sterile conditions, and based on these tests genus Enterococcus (S203) Thirty-eight percent, Bacillus (S202) eighteen percent, Pseudomonas (S200) 17.6 percent and lactobacilli (S199), 26.7 percent of all isolates were included.
3.2. The Results of the Evaluation of the Probiotic Properties
- -Resistance to acids and bile saltsThe isolated bacteria were able to tolerate acidic conditions at different times. As well as the different concentrations of bacteria were resistant to bile salts which resistance patterns to any of the conditions are shown in figures 1 and 2.
 | Figure 1. Resistance Pattern to acidic conditions in selected bacteria |
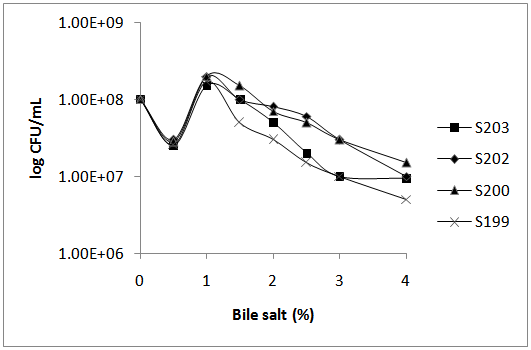 | Figure 2. Resistance Pattern to bile salts in selected bacteria |
3.3. Results of the Assessment of Isolated Probiotics Effect on Prawn Growth
- Among the four probiotic isolates, mixtreof genera Bacillus and Enterococcus plus prawn food (50% mixture of two bacteria plus 50% of prawnture of food) that were pelleted and the amount of 2% by weight of prawnand 3-4 times a day were added to each aquarium, showed the greatest impact on the life of prawn and biometric parameters.Analysis of data variance resulted from a completely randomized program and using the SAS version 9.1, GLM procedure and comparing the mean with Duncan test at 5% probability level was performed.The results show that between the control treatment and probiotic during the breeding (15 and 45 days) about the growth parameters including the daily growth average , body weight gain, increased body length, increased carapace length and SGR and Survival percentage, there was a statistical difference (P <0.05).Also for the total length, carapace length and weight between the control treatment and probiotics in prawn breeding periods (15 and 45 days) at the 5% probability level, there was a statistical difference (P <0.05) - ( Figure 4).During the period, an increase in weight of prawns fed with diets containing probiotics was seen (Figure 3) and this increase in probiotic treatments with a control treatment was significantly different (P <0.05).At the end of the breeding period (day 45) the highest and lowest values prawn were observed, in treatments containing probiotics respectively (Fig. 3).
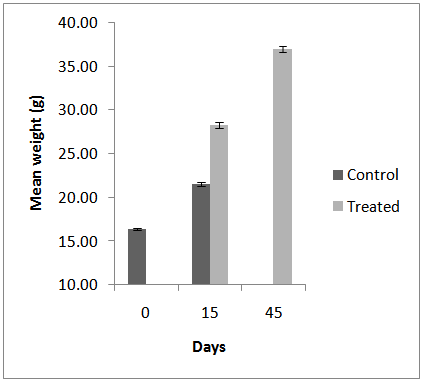 | Figure 3. Effect of the control and probiotic treatments during the 15-45 days breeding periods on prawns' weight |
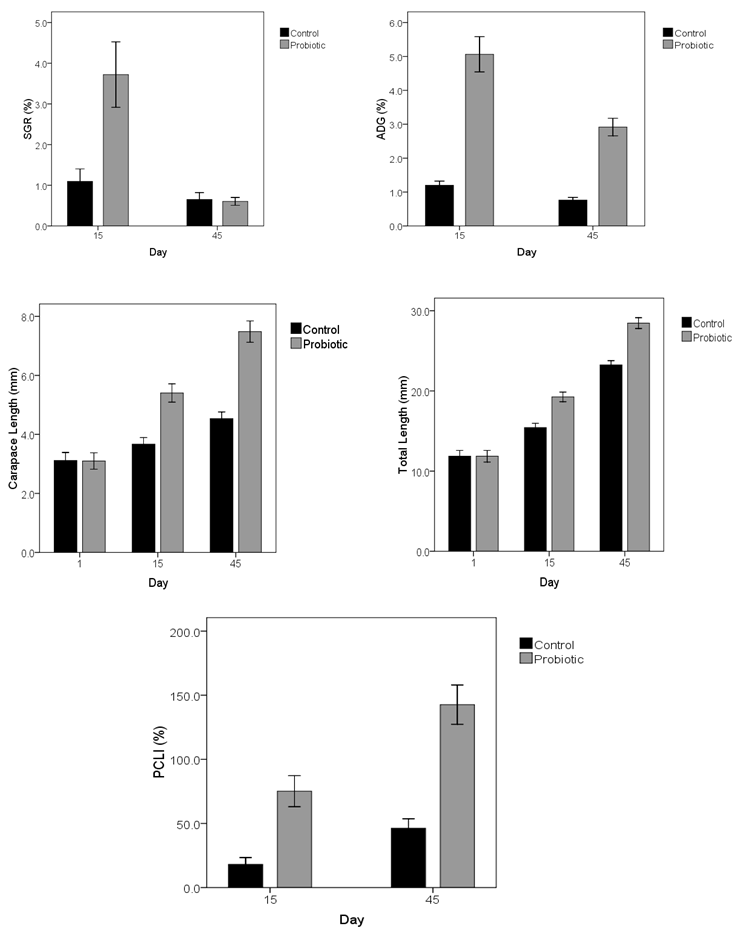 | Figure 4. Effect of control and probiotic treatments in 15 and 45 days of breeding periods on growth parameters of prawn |
3.4. Identification Results of Probiotics by Sequencing 16SrRNA
- Based on gene homology and analysis through BLAST (Blastn algorithm) on the site http:// www.ncbi.nih.gov /, bp500 sequences of isolated probiotics showed that S202 and S203 bacteria, 99% were owned by Bacillus licheniformis and Enterococcus faecium that by access numbers of JQ388689 and JQ619997 and names of Bacillus licheniformis strain RK202 and Enterococcus faecium strain RK 203 according to Table 1 were recorded in the NCBI database.
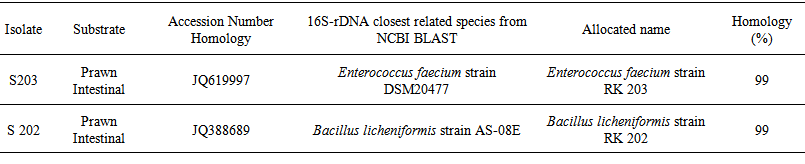 | Table 1. Identification of probiotics through analysis of 16SrDNA |
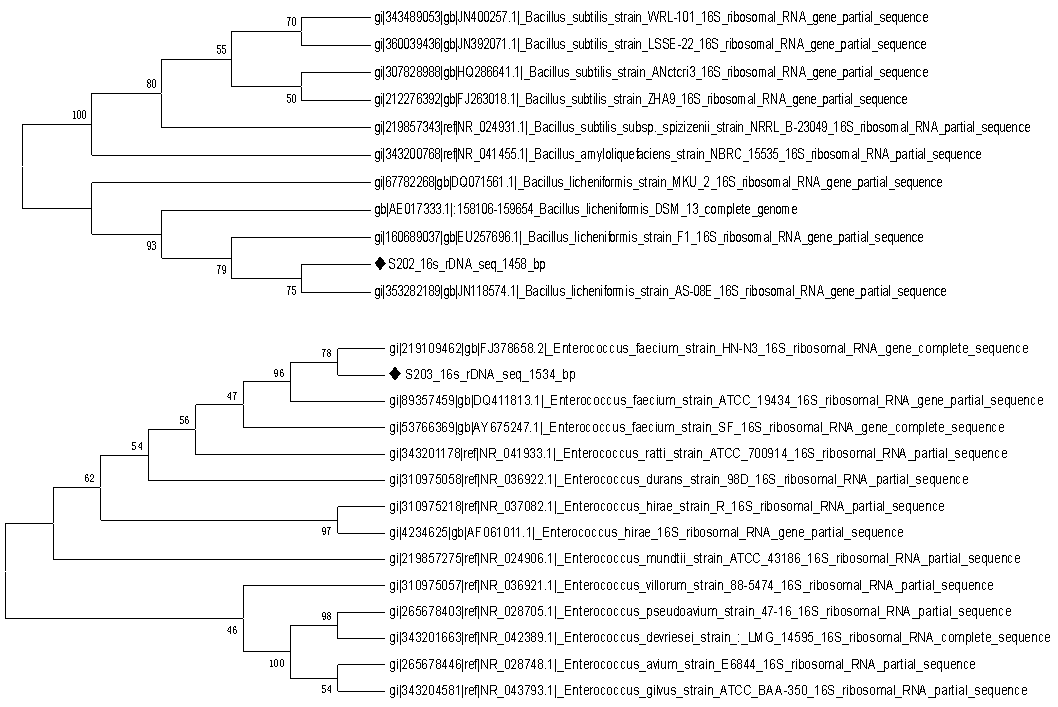 | Figure 6. Phylogenetic tree of Two strains RK 202 and 203 RK based on sequence analysis of 16S ribosomal RNA (isolated probiotics is indicated by sign) |
4. Discussion and Conclusions
- Nowadays, the use of probiotics in nutrition, disease control, an increase in aquaculture production, reducing antibiotic resistance and their use as an adjuvant to increase the immune response in fish is important in aquaculture. Since Macrobrachium rosenbergii due to taste of meat, low in cholesterol and high in protein reserves commercially is important.Usage of a way that in addition to being cost-efficient causes to increase this aquatic production is important that the route is paved through consumption of probioticsplus food.In this study, after isolation and Gram staining and differentiation tests, the influence of probiotics on prawn biometric parameters were studied and effective probiotics on growth, 16s rRNA were identified by the technique.In this study, four genera S202, S203, S199, S200 regarding their probiotic properties, were evaluated. Every 4 genera are resistant to pH 2-4 and bile salts and among 4 genera, S202 and S203, have the greatest ability to withstand acidic conditions and resistance to bile that the research methods were same as Anil K. Patel and colleagues in 2009 [6].Aeromonas and Vibrio species are responsible for the majority of disease and mortality in the prawn aquaculture [15].S202 and S203 probiotics have the greatest anti-microbial ability due to produce hydrogen peroxide, bacteriocins and lactic acid were against pathogens.Such that by disabling Hydrogen peroxide by catalase, and alkalinize the solution obtained from probiotics, these bacteria also lost their antimicrobial properties. These reviews were consistent with the researches of Aslim and colleagues, who in 2005 took the probiotic to control pathogens [19].S202 (of Bacillus licheniformis genus) and S203 (of Enterococcus faecium genus) strains were susceptible than most of the studied antibiotics.This property is an important component of the probiotic properties must be qualified because the presence of plasmid genes of antibiotic resistance through the mechanism of among the resistant bacteria through the conjugation mechanism will be transferred between the intestinal flora bacteria [19].In order that a probiotic can have a nutritional role must be able to bind to intestinal cells. In this study, each of the four different levels of bacteria was attached to intestinal cells and between them strains S202 and S203 showed higher binding ability [11]Given that each of the bacteria, S202, S203, S199 and S200 showed us a different probiotic properties, the selection of appropriate probiotics on the basis of the positive impact of these bacteria on the biometric indicators of prawns such as percentage of body weight, percentage of carapace length, percentage of the total length of the prawn body took place .Among these bacteria, S202 and S203 strains mixture with prawn food (50 wt% mixture of two bacteria with 50% prawn food) that were pelleted and the amount of 2% by weight of prawn and 3-4 times a day were added to each aquarium, has the greatest impact on biometric indices of prawns. These ingredients can be used to produce the fish in bulk.After the effect of bacteria on the growth and survival of prawn, genus and species of these bacteria were determined by analysis of 16S rRNA genes [9-16]Based on gene homology and analysis using the software BLAST (Blastn algorithm) on the site http://www. ncbi.nih.gov /, it was shown that bacteria, S202 and S203, respectively, belong to the 99% rate to species of Enterococcus faecium and Bacillus licheniformis and were recorded in the NCBI database by names of Bacillus licheniformis strain RK202 and Enterococcus faecium strain RK 203.
 Abstract
Abstract Reference
Reference Full-Text PDF
Full-Text PDF Full-text HTML
Full-text HTML